Aryan Mobiny
Few Is Enough: Task-Augmented Active Meta-Learning for Brain Cell Classification
Jul 09, 2020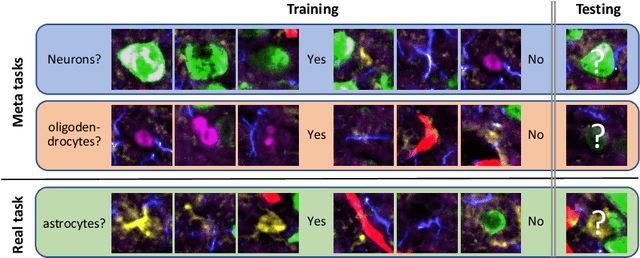
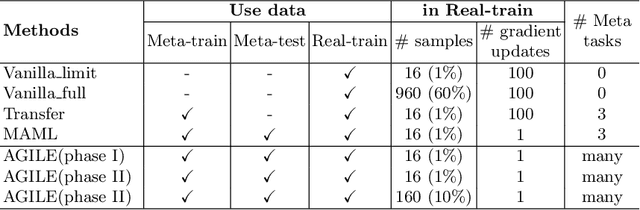
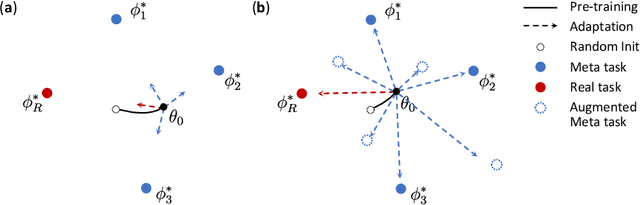
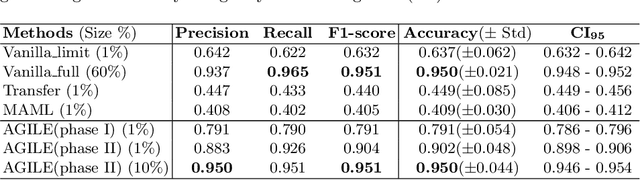
Abstract:Deep Neural Networks (or DNNs) must constantly cope with distribution changes in the input data when the task of interest or the data collection protocol changes. Retraining a network from scratch to combat this issue poses a significant cost. Meta-learning aims to deliver an adaptive model that is sensitive to these underlying distribution changes, but requires many tasks during the meta-training process. In this paper, we propose a tAsk-auGmented actIve meta-LEarning (AGILE) method to efficiently adapt DNNs to new tasks by using a small number of training examples. AGILE combines a meta-learning algorithm with a novel task augmentation technique which we use to generate an initial adaptive model. It then uses Bayesian dropout uncertainty estimates to actively select the most difficult samples when updating the model to a new task. This allows AGILE to learn with fewer tasks and a few informative samples, achieving high performance with a limited dataset. We perform our experiments using the brain cell classification task and compare the results to a plain meta-learning model trained from scratch. We show that the proposed task-augmented meta-learning framework can learn to classify new cell types after a single gradient step with a limited number of training samples. We show that active learning with Bayesian uncertainty can further improve the performance when the number of training samples is extremely small. Using only 1% of the training data and a single update step, we achieved 90% accuracy on the new cell type classification task, a 50% points improvement over a state-of-the-art meta-learning algorithm.
StyPath: Style-Transfer Data Augmentation For Robust Histology Image Classification
Jul 09, 2020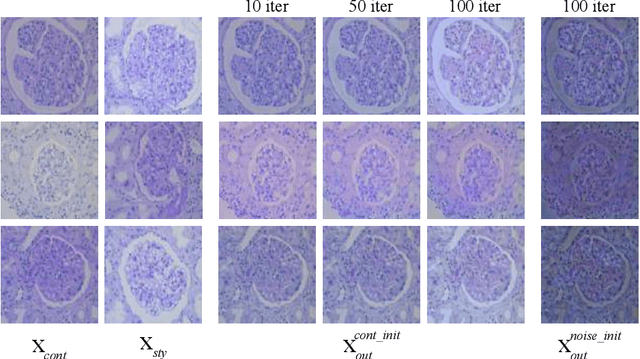
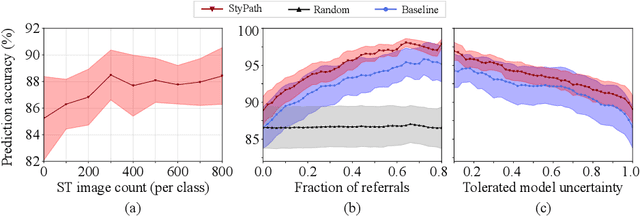
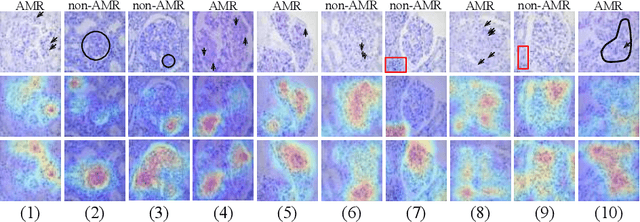
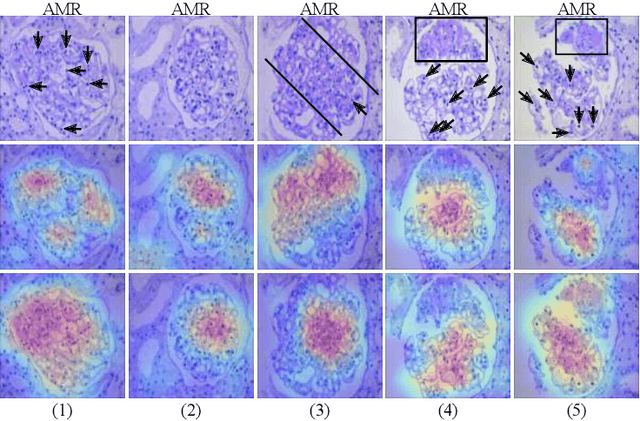
Abstract:The classification of Antibody Mediated Rejection (AMR) in kidney transplant remains challenging even for experienced nephropathologists; this is partly because histological tissue stain analysis is often characterized by low inter-observer agreement and poor reproducibility. One of the implicated causes for inter-observer disagreement is the variability of tissue stain quality between (and within) pathology labs, coupled with the gradual fading of archival sections. Variations in stain colors and intensities can make tissue evaluation difficult for pathologists, ultimately affecting their ability to describe relevant morphological features. Being able to accurately predict the AMR status based on kidney histology images is crucial for improving patient treatment and care. We propose a novel pipeline to build robust deep neural networks for AMR classification based on StyPath, a histological data augmentation technique that leverages a light weight style-transfer algorithm as a means to reduce sample-specific bias. Each image was generated in 1.84 +- 0.03 seconds using a single GTX TITAN V gpu and pytorch, making it faster than other popular histological data augmentation techniques. We evaluated our model using a Monte Carlo (MC) estimate of Bayesian performance and generate an epistemic measure of uncertainty to compare both the baseline and StyPath augmented models. We also generated Grad-CAM representations of the results which were assessed by an experienced nephropathologist; we used this qualitative analysis to elucidate on the assumptions being made by each model. Our results imply that our style-transfer augmentation technique improves histological classification performance (reducing error from 14.8% to 11.5%) and generalization ability.
DECAPS: Detail-Oriented Capsule Networks
Jul 09, 2020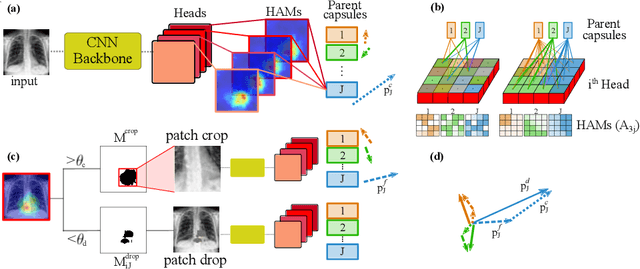
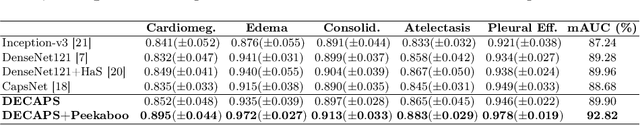
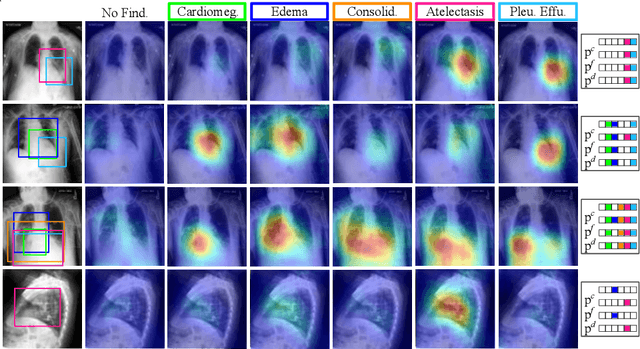
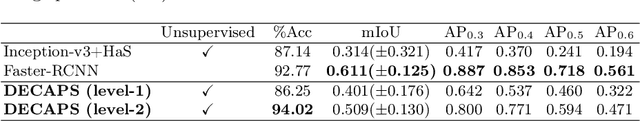
Abstract:Capsule Networks (CapsNets) have demonstrated to be a promising alternative to Convolutional Neural Networks (CNNs). However, they often fall short of state-of-the-art accuracies on large-scale high-dimensional datasets. We propose a Detail-Oriented Capsule Network (DECAPS) that combines the strength of CapsNets with several novel techniques to boost its classification accuracies. First, DECAPS uses an Inverted Dynamic Routing (IDR) mechanism to group lower-level capsules into heads before sending them to higher-level capsules. This strategy enables capsules to selectively attend to small but informative details within the data which may be lost during pooling operations in CNNs. Second, DECAPS employs a Peekaboo training procedure, which encourages the network to focus on fine-grained information through a second-level attention scheme. Finally, the distillation process improves the robustness of DECAPS by averaging over the original and attended image region predictions. We provide extensive experiments on the CheXpert and RSNA Pneumonia datasets to validate the effectiveness of DECAPS. Our networks achieve state-of-the-art accuracies not only in classification (increasing the average area under ROC curves from 87.24% to 92.82% on the CheXpert dataset) but also in the weakly-supervised localization of diseased areas (increasing average precision from 41.7% to 80% for the RSNA Pneumonia detection dataset).
Radiologist-Level COVID-19 Detection Using CT Scans with Detail-Oriented Capsule Networks
Apr 16, 2020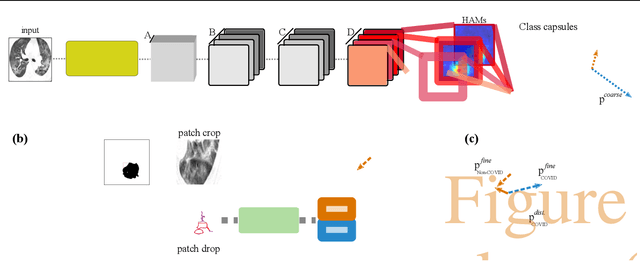
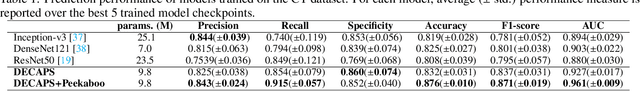
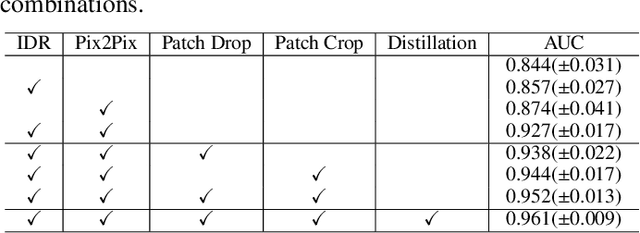
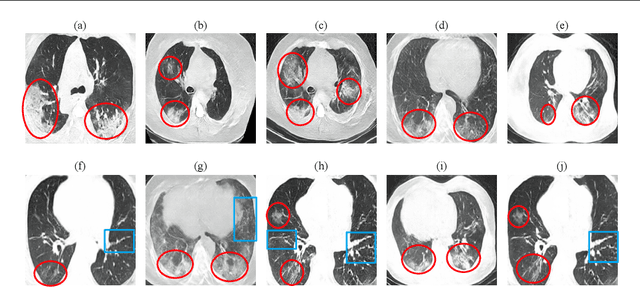
Abstract:Radiographic images offer an alternative method for the rapid screening and monitoring of Coronavirus Disease 2019 (COVID-19) patients. This approach is limited by the shortage of radiology experts who can provide a timely interpretation of these images. Motivated by this challenge, our paper proposes a novel learning architecture, called Detail-Oriented Capsule Networks (DECAPS), for the automatic diagnosis of COVID-19 from Computed Tomography (CT) scans. Our network combines the strength of Capsule Networks with several architecture improvements meant to boost classification accuracies. First, DECAPS uses an Inverted Dynamic Routing mechanism which increases model stability by preventing the passage of information from non-descriptive regions. Second, DECAPS employs a Peekaboo training procedure which uses a two-stage patch crop and drop strategy to encourage the network to generate activation maps for every target concept. The network then uses the activation maps to focus on regions of interest and combines both coarse and fine-grained representations of the data. Finally, we use a data augmentation method based on conditional generative adversarial networks to deal with the issue of data scarcity. Our model achieves 84.3% precision, 91.5% recall, and 96.1% area under the ROC curve, significantly outperforming state-of-the-art methods. We compare the performance of the DECAPS model with three experienced, well-trained thoracic radiologists and show that the architecture significantly outperforms them. While further studies on larger datasets are required to confirm this finding, our results imply that architectures like DECAPS can be used to assist radiologists in the CT scan mediated diagnosis of COVID-19.
DVNet: A Memory-Efficient Three-Dimensional CNN for Large-Scale Neurovascular Reconstruction
Feb 04, 2020
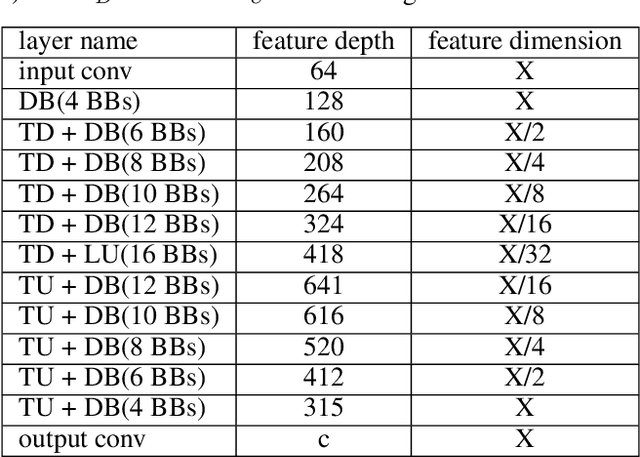


Abstract:Maps of brain microarchitecture are important for understanding neurological function and behavior, including alterations caused by chronic conditions such as neurodegenerative disease. Techniques such as knife-edge scanning microscopy (KESM) provide the potential for whole organ imaging at sub-cellular resolution. However, multi-terabyte data sizes make manual annotation impractical and automatic segmentation challenging. Densely packed cells combined with interconnected microvascular networks are a challenge for current segmentation algorithms. The massive size of high-throughput microscopy data necessitates fast and largely unsupervised algorithms. In this paper, we investigate a fully-convolutional, deep, and densely-connected encoder-decoder for pixel-wise semantic segmentation. The excessive memory complexity often encountered with deep and dense networks is mitigated using skip connections, resulting in fewer parameters and enabling a significant performance increase over prior architectures. The proposed network provides superior performance for semantic segmentation problems applied to open-source benchmarks. We finally demonstrate our network for cellular and microvascular segmentation, enabling quantitative metrics for organ-scale neurovascular analysis.
DropConnect Is Effective in Modeling Uncertainty of Bayesian Deep Networks
Jun 07, 2019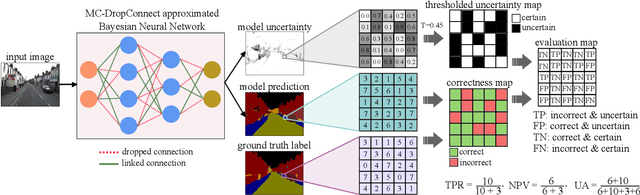
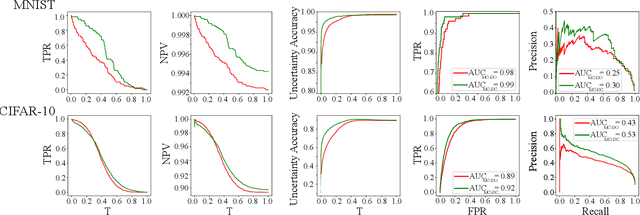
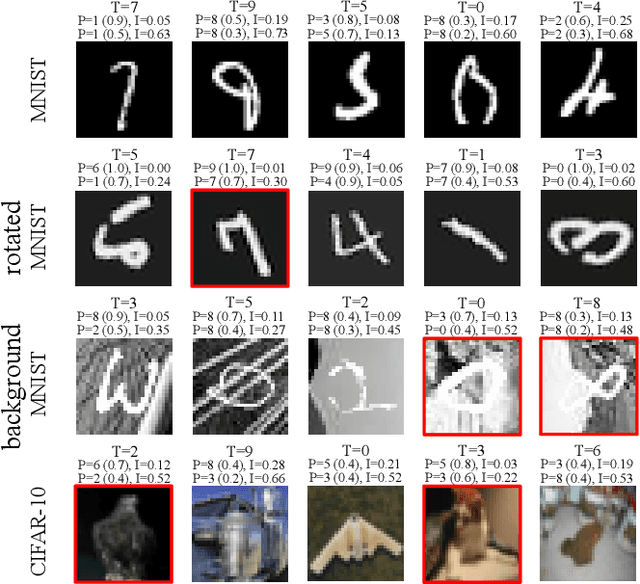
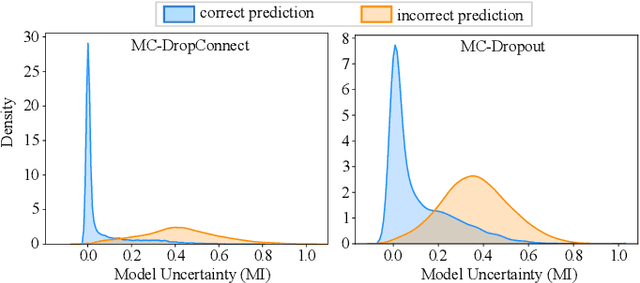
Abstract:Deep neural networks (DNNs) have achieved state-of-the-art performances in many important domains, including medical diagnosis, security, and autonomous driving. In these domains where safety is highly critical, an erroneous decision can result in serious consequences. While a perfect prediction accuracy is not always achievable, recent work on Bayesian deep networks shows that it is possible to know when DNNs are more likely to make mistakes. Knowing what DNNs do not know is desirable to increase the safety of deep learning technology in sensitive applications. Bayesian neural networks attempt to address this challenge. However, traditional approaches are computationally intractable and do not scale well to large, complex neural network architectures. In this paper, we develop a theoretical framework to approximate Bayesian inference for DNNs by imposing a Bernoulli distribution on the model weights. This method, called MC-DropConnect, gives us a tool to represent the model uncertainty with little change in the overall model structure or computational cost. We extensively validate the proposed algorithm on multiple network architectures and datasets for classification and semantic segmentation tasks. We also propose new metrics to quantify the uncertainty estimates. This enables an objective comparison between MC-DropConnect and prior approaches. Our empirical results demonstrate that the proposed framework yields significant improvement in both prediction accuracy and uncertainty estimation quality compared to the state of the art.
Lung Cancer Screening Using Adaptive Memory-Augmented Recurrent Networks
Sep 07, 2018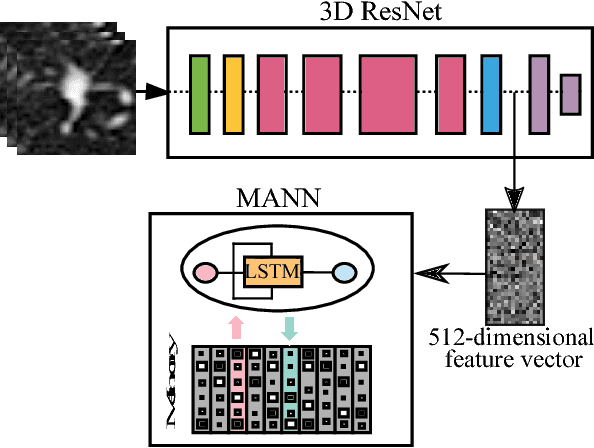
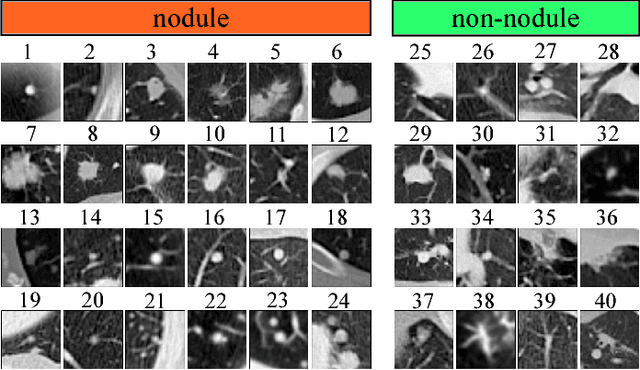
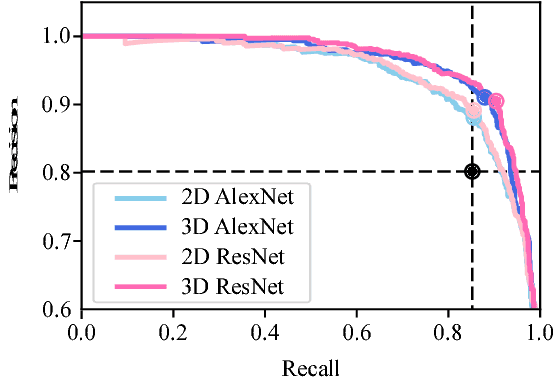
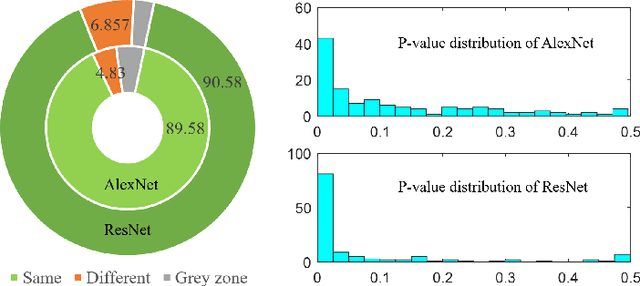
Abstract:In this paper, we investigate the effectiveness of deep learning techniques for lung nodule classification in computed tomography scans. Using less than 10,000 training examples, our deep networks perform two times better than a standard radiology software. Visualization of the networks' neurons reveals semantically meaningful features that are consistent with the clinical knowledge and radiologists' perception. Our paper also proposes a novel framework for rapidly adapting deep networks to the radiologists' feedback, or change in the data due to the shift in sensor's resolution or patient population. The classification accuracy of our approach remains above 80% while popular deep networks' accuracy is around chance. Finally, we provide in-depth analysis of our framework by asking a radiologist to examine important networks' features and perform blind re-labeling of networks' mistakes.
Text-Independent Speaker Verification Using Long Short-Term Memory Networks
Sep 07, 2018
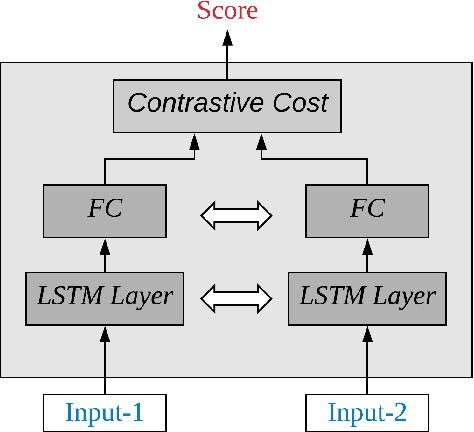
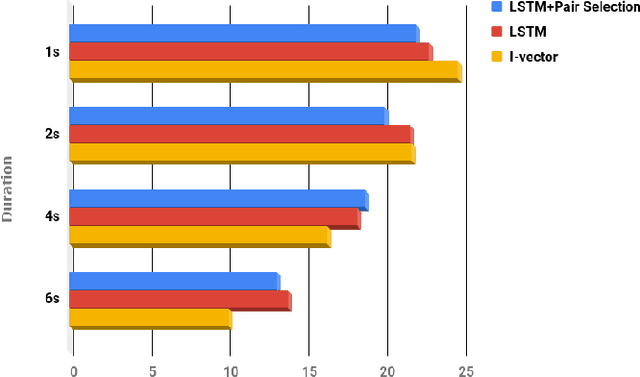
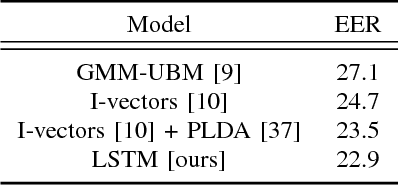
Abstract:In this paper, an architecture based on Long Short-Term Memory Networks has been proposed for the text-independent scenario which is aimed to capture the temporal speaker-related information by operating over traditional speech features. For speaker verification, at first, a background model must be created for speaker representation. Then, in enrollment stage, the speaker models will be created based on the enrollment utterances. For this work, the model will be trained in an end-to-end fashion to combine the first two stages. The main goal of end-to-end training is the model being optimized to be consistent with the speaker verification protocol. The end- to-end training jointly learns the background and speaker models by creating the representation space. The LSTM architecture is trained to create a discrimination space for validating the match and non-match pairs for speaker verification. The proposed architecture demonstrate its superiority in the text-independent compared to other traditional methods.
Fast CapsNet for Lung Cancer Screening
Jun 19, 2018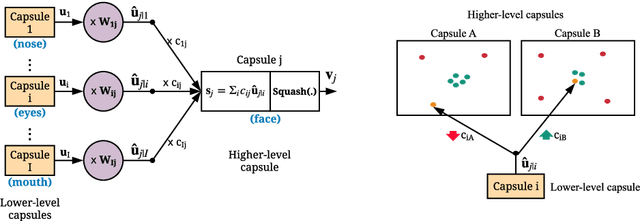
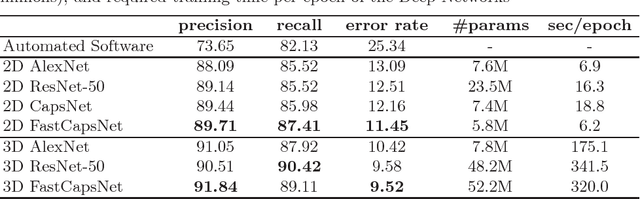
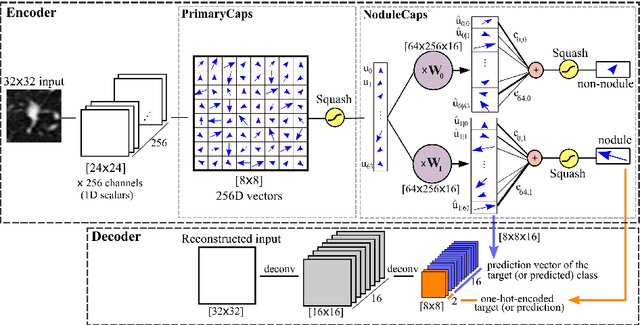
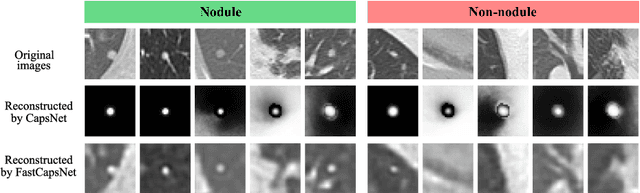
Abstract:Lung cancer is the leading cause of cancer-related deaths in the past several years. A major challenge in lung cancer screening is the detection of lung nodules from computed tomography (CT) scans. State-of-the-art approaches in automated lung nodule classification use deep convolutional neural networks (CNNs). However, these networks require a large number of training samples to generalize well. This paper investigates the use of capsule networks (CapsNets) as an alternative to CNNs. We show that CapsNets significantly outperforms CNNs when the number of training samples is small. To increase the computational efficiency, our paper proposes a consistent dynamic routing mechanism that results in $3\times$ speedup of CapsNet. Finally, we show that the original image reconstruction method of CapNets performs poorly on lung nodule data. We propose an efficient alternative, called convolutional decoder, that yields lower reconstruction error and higher classification accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge