Pietro Antonio Cicalese
Few Is Enough: Task-Augmented Active Meta-Learning for Brain Cell Classification
Jul 09, 2020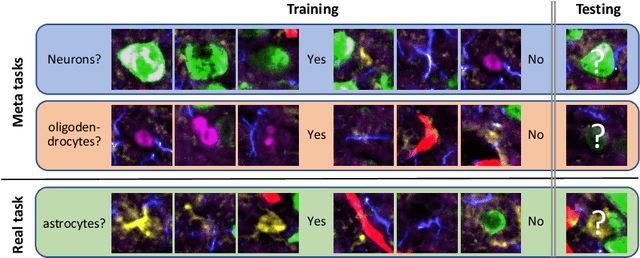
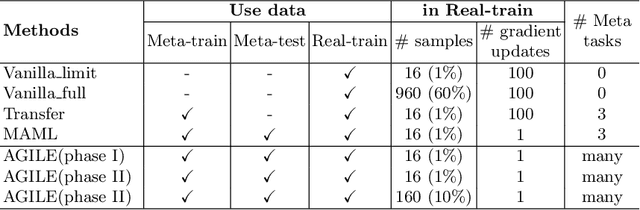
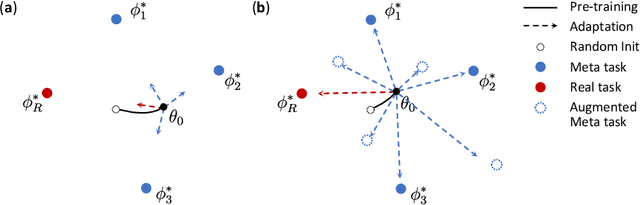
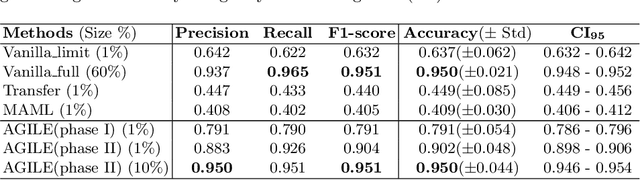
Abstract:Deep Neural Networks (or DNNs) must constantly cope with distribution changes in the input data when the task of interest or the data collection protocol changes. Retraining a network from scratch to combat this issue poses a significant cost. Meta-learning aims to deliver an adaptive model that is sensitive to these underlying distribution changes, but requires many tasks during the meta-training process. In this paper, we propose a tAsk-auGmented actIve meta-LEarning (AGILE) method to efficiently adapt DNNs to new tasks by using a small number of training examples. AGILE combines a meta-learning algorithm with a novel task augmentation technique which we use to generate an initial adaptive model. It then uses Bayesian dropout uncertainty estimates to actively select the most difficult samples when updating the model to a new task. This allows AGILE to learn with fewer tasks and a few informative samples, achieving high performance with a limited dataset. We perform our experiments using the brain cell classification task and compare the results to a plain meta-learning model trained from scratch. We show that the proposed task-augmented meta-learning framework can learn to classify new cell types after a single gradient step with a limited number of training samples. We show that active learning with Bayesian uncertainty can further improve the performance when the number of training samples is extremely small. Using only 1% of the training data and a single update step, we achieved 90% accuracy on the new cell type classification task, a 50% points improvement over a state-of-the-art meta-learning algorithm.
StyPath: Style-Transfer Data Augmentation For Robust Histology Image Classification
Jul 09, 2020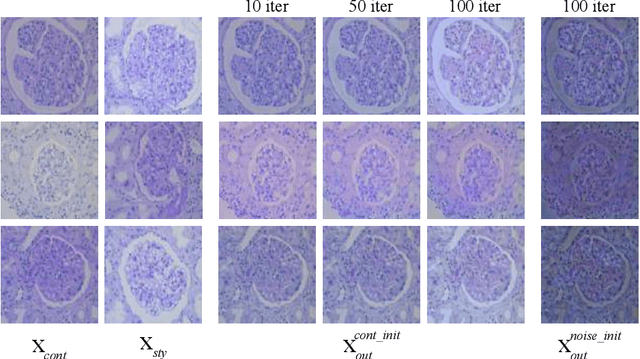
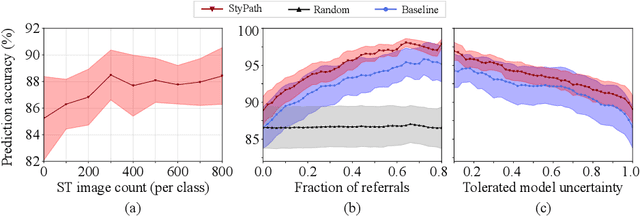
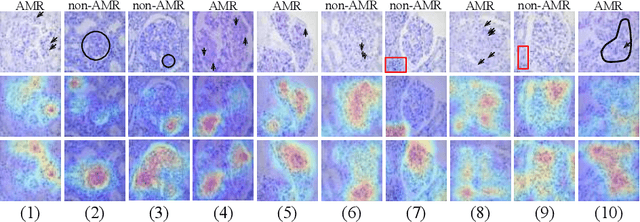
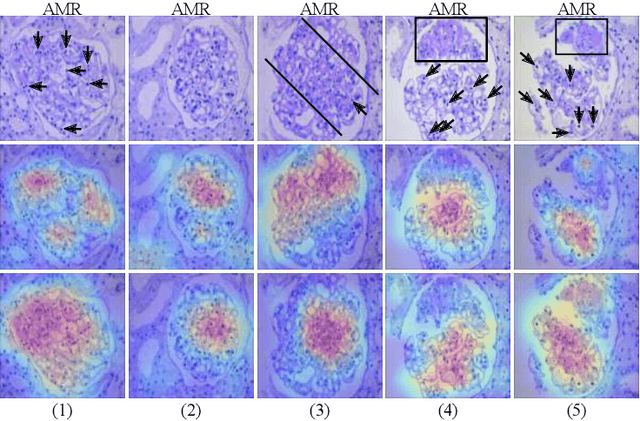
Abstract:The classification of Antibody Mediated Rejection (AMR) in kidney transplant remains challenging even for experienced nephropathologists; this is partly because histological tissue stain analysis is often characterized by low inter-observer agreement and poor reproducibility. One of the implicated causes for inter-observer disagreement is the variability of tissue stain quality between (and within) pathology labs, coupled with the gradual fading of archival sections. Variations in stain colors and intensities can make tissue evaluation difficult for pathologists, ultimately affecting their ability to describe relevant morphological features. Being able to accurately predict the AMR status based on kidney histology images is crucial for improving patient treatment and care. We propose a novel pipeline to build robust deep neural networks for AMR classification based on StyPath, a histological data augmentation technique that leverages a light weight style-transfer algorithm as a means to reduce sample-specific bias. Each image was generated in 1.84 +- 0.03 seconds using a single GTX TITAN V gpu and pytorch, making it faster than other popular histological data augmentation techniques. We evaluated our model using a Monte Carlo (MC) estimate of Bayesian performance and generate an epistemic measure of uncertainty to compare both the baseline and StyPath augmented models. We also generated Grad-CAM representations of the results which were assessed by an experienced nephropathologist; we used this qualitative analysis to elucidate on the assumptions being made by each model. Our results imply that our style-transfer augmentation technique improves histological classification performance (reducing error from 14.8% to 11.5%) and generalization ability.
DECAPS: Detail-Oriented Capsule Networks
Jul 09, 2020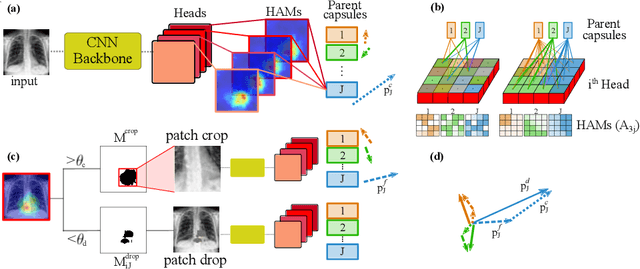
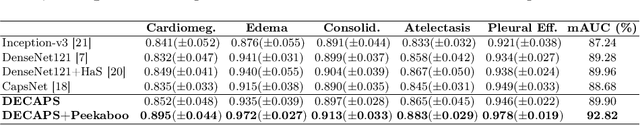
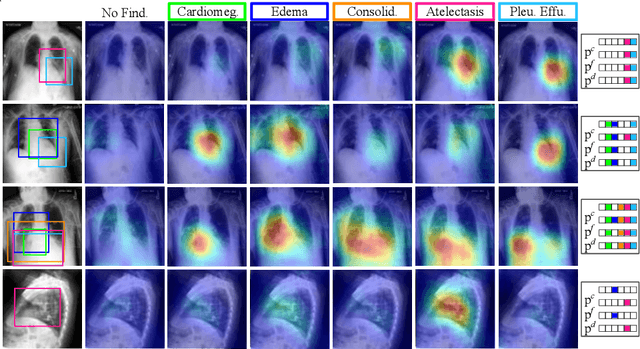
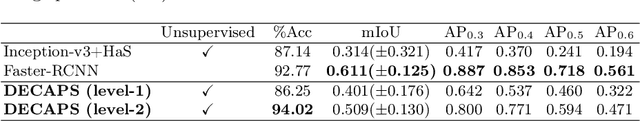
Abstract:Capsule Networks (CapsNets) have demonstrated to be a promising alternative to Convolutional Neural Networks (CNNs). However, they often fall short of state-of-the-art accuracies on large-scale high-dimensional datasets. We propose a Detail-Oriented Capsule Network (DECAPS) that combines the strength of CapsNets with several novel techniques to boost its classification accuracies. First, DECAPS uses an Inverted Dynamic Routing (IDR) mechanism to group lower-level capsules into heads before sending them to higher-level capsules. This strategy enables capsules to selectively attend to small but informative details within the data which may be lost during pooling operations in CNNs. Second, DECAPS employs a Peekaboo training procedure, which encourages the network to focus on fine-grained information through a second-level attention scheme. Finally, the distillation process improves the robustness of DECAPS by averaging over the original and attended image region predictions. We provide extensive experiments on the CheXpert and RSNA Pneumonia datasets to validate the effectiveness of DECAPS. Our networks achieve state-of-the-art accuracies not only in classification (increasing the average area under ROC curves from 87.24% to 92.82% on the CheXpert dataset) but also in the weakly-supervised localization of diseased areas (increasing average precision from 41.7% to 80% for the RSNA Pneumonia detection dataset).
Radiologist-Level COVID-19 Detection Using CT Scans with Detail-Oriented Capsule Networks
Apr 16, 2020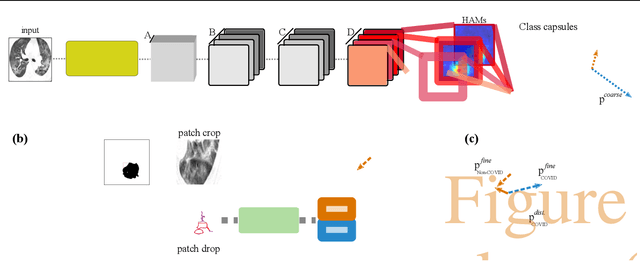
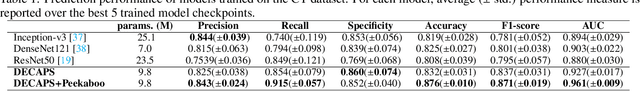
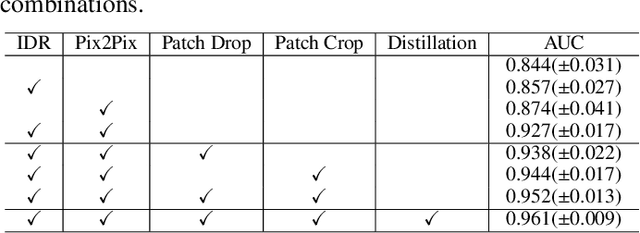
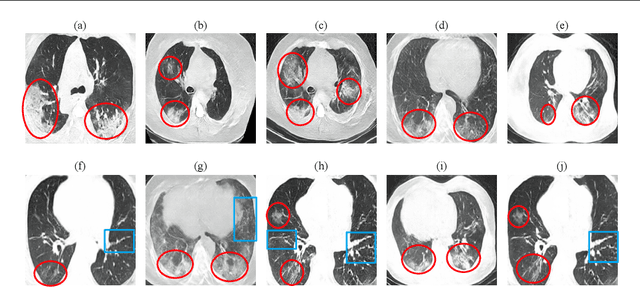
Abstract:Radiographic images offer an alternative method for the rapid screening and monitoring of Coronavirus Disease 2019 (COVID-19) patients. This approach is limited by the shortage of radiology experts who can provide a timely interpretation of these images. Motivated by this challenge, our paper proposes a novel learning architecture, called Detail-Oriented Capsule Networks (DECAPS), for the automatic diagnosis of COVID-19 from Computed Tomography (CT) scans. Our network combines the strength of Capsule Networks with several architecture improvements meant to boost classification accuracies. First, DECAPS uses an Inverted Dynamic Routing mechanism which increases model stability by preventing the passage of information from non-descriptive regions. Second, DECAPS employs a Peekaboo training procedure which uses a two-stage patch crop and drop strategy to encourage the network to generate activation maps for every target concept. The network then uses the activation maps to focus on regions of interest and combines both coarse and fine-grained representations of the data. Finally, we use a data augmentation method based on conditional generative adversarial networks to deal with the issue of data scarcity. Our model achieves 84.3% precision, 91.5% recall, and 96.1% area under the ROC curve, significantly outperforming state-of-the-art methods. We compare the performance of the DECAPS model with three experienced, well-trained thoracic radiologists and show that the architecture significantly outperforms them. While further studies on larger datasets are required to confirm this finding, our results imply that architectures like DECAPS can be used to assist radiologists in the CT scan mediated diagnosis of COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge