Artem Razumov
The state-of-the-art in Cardiac MRI Reconstruction: Results of the CMRxRecon Challenge in MICCAI 2023
Apr 01, 2024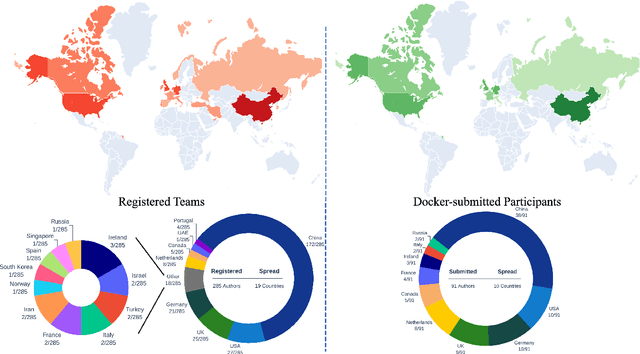
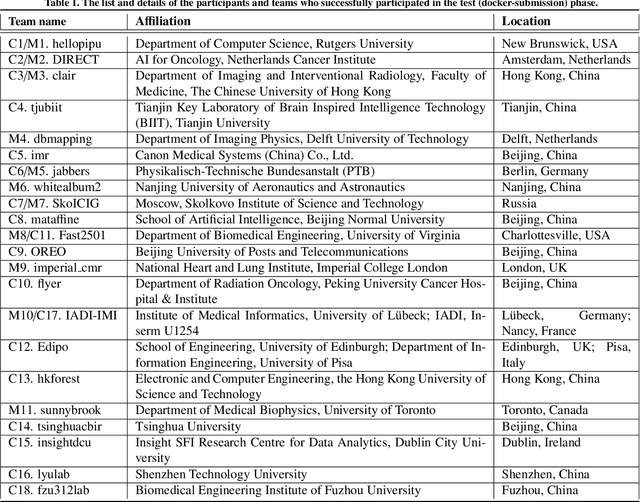
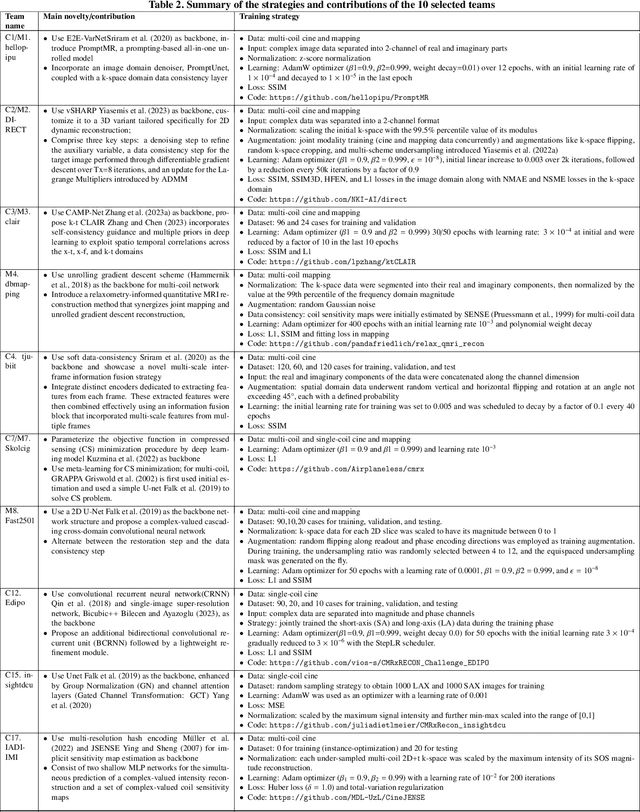
Abstract:Cardiac MRI, crucial for evaluating heart structure and function, faces limitations like slow imaging and motion artifacts. Undersampling reconstruction, especially data-driven algorithms, has emerged as a promising solution to accelerate scans and enhance imaging performance using highly under-sampled data. Nevertheless, the scarcity of publicly available cardiac k-space datasets and evaluation platform hinder the development of data-driven reconstruction algorithms. To address this issue, we organized the Cardiac MRI Reconstruction Challenge (CMRxRecon) in 2023, in collaboration with the 26th International Conference on MICCAI. CMRxRecon presented an extensive k-space dataset comprising cine and mapping raw data, accompanied by detailed annotations of cardiac anatomical structures. With overwhelming participation, the challenge attracted more than 285 teams and over 600 participants. Among them, 22 teams successfully submitted Docker containers for the testing phase, with 7 teams submitted for both cine and mapping tasks. All teams use deep learning based approaches, indicating that deep learning has predominately become a promising solution for the problem. The first-place winner of both tasks utilizes the E2E-VarNet architecture as backbones. In contrast, U-Net is still the most popular backbone for both multi-coil and single-coil reconstructions. This paper provides a comprehensive overview of the challenge design, presents a summary of the submitted results, reviews the employed methods, and offers an in-depth discussion that aims to inspire future advancements in cardiac MRI reconstruction models. The summary emphasizes the effective strategies observed in Cardiac MRI reconstruction, including backbone architecture, loss function, pre-processing techniques, physical modeling, and model complexity, thereby providing valuable insights for further developments in this field.
Autofocusing+: Noise-Resilient Motion Correction in Magnetic Resonance Imaging
Mar 10, 2022



Abstract:Image corruption by motion artifacts is an ingrained problem in Magnetic Resonance Imaging (MRI). In this work, we propose a neural network-based regularization term to enhance Autofocusing, a classic optimization-based method to remove motion artifacts. The method takes the best of both worlds: the optimization-based routine iteratively executes the blind demotion and deep learning-based prior penalizes for unrealistic restorations and speeds up the convergence. We validate the method on three models of motion trajectories, using synthetic and real noisy data. The method proves resilient to noise and anatomic structure variation, outperforming the state-of-the-art demotion methods.
Optimal MRI Undersampling Patterns for Ultimate Benefit of Medical Vision Tasks
Aug 10, 2021



Abstract:To accelerate MRI, the field of compressed sensing is traditionally concerned with optimizing the image quality after a partial undersampling of the measurable $\textit{k}$-space. In our work, we propose to change the focus from the quality of the reconstructed image to the quality of the downstream image analysis outcome. Specifically, we propose to optimize the patterns according to how well a sought-after pathology could be detected or localized in the reconstructed images. We find the optimal undersampling patterns in $\textit{k}$-space that maximize target value functions of interest in commonplace medical vision problems (reconstruction, segmentation, and classification) and propose a new iterative gradient sampling routine universally suitable for these tasks. We validate the proposed MRI acceleration paradigm on three classical medical datasets, demonstrating a noticeable improvement of the target metrics at the high acceleration factors (for the segmentation problem at $\times$16 acceleration, we report up to 12% improvement in Dice score over the other undersampling patterns).
Comparison of UNet, ENet, and BoxENet for Segmentation of Mast Cells in Scans of Histological Slices
Oct 15, 2019
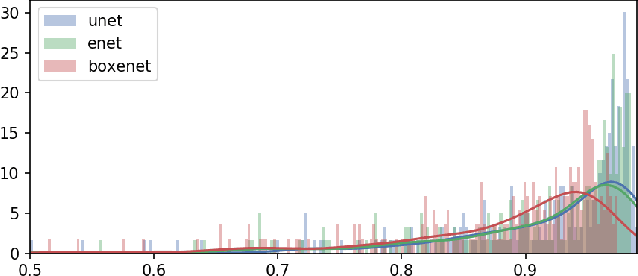
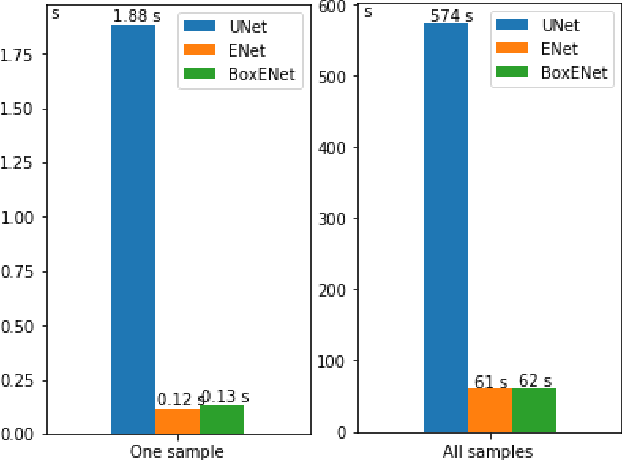

Abstract:Deep neural networks show high accuracy in theproblem of semantic and instance segmentation of biomedicaldata. However, this approach is computationally expensive. Thecomputational cost may be reduced with network simplificationafter training or choosing the proper architecture, which providessegmentation with less accuracy but does it much faster. In thepresent study, we analyzed the accuracy and performance ofUNet and ENet architectures for the problem of semantic imagesegmentation. In addition, we investigated the ENet architecture by replacing of some convolution layers with box-convolutionlayers. The analysis performed on the original dataset consisted of histology slices with mast cells. These cells provide a region forsegmentation with different types of borders, which vary fromclearly visible to ragged. ENet was less accurate than UNet byonly about 1-2%, but ENet performance was 8-15 times faster than UNet one.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge