Konstantin Ushenin
$ abla^2$DFT: A Universal Quantum Chemistry Dataset of Drug-Like Molecules and a Benchmark for Neural Network Potentials
Jun 20, 2024Abstract:Methods of computational quantum chemistry provide accurate approximations of molecular properties crucial for computer-aided drug discovery and other areas of chemical science. However, high computational complexity limits the scalability of their applications. Neural network potentials (NNPs) are a promising alternative to quantum chemistry methods, but they require large and diverse datasets for training. This work presents a new dataset and benchmark called $\nabla^2$DFT that is based on the nablaDFT. It contains twice as much molecular structures, three times more conformations, new data types and tasks, and state-of-the-art models. The dataset includes energies, forces, 17 molecular properties, Hamiltonian and overlap matrices, and a wavefunction object. All calculations were performed at the DFT level ($\omega$B97X-D/def2-SVP) for each conformation. Moreover, $\nabla^2$DFT is the first dataset that contains relaxation trajectories for a substantial number of drug-like molecules. We also introduce a novel benchmark for evaluating NNPs in molecular property prediction, Hamiltonian prediction, and conformational optimization tasks. Finally, we propose an extendable framework for training NNPs and implement 10 models within it.
Benefits of mirror weight symmetry for 3D mesh segmentation in biomedical applications
Sep 29, 2023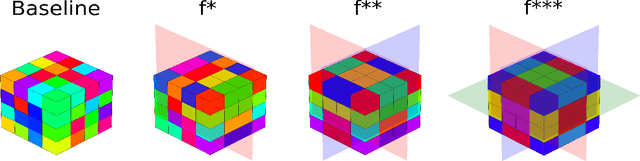
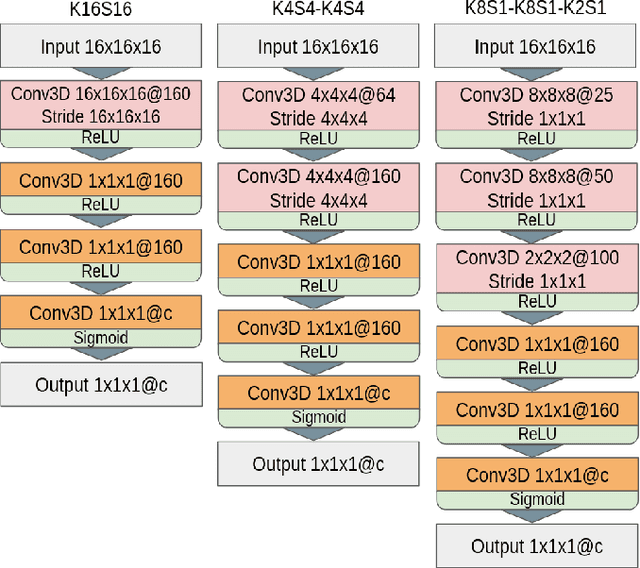
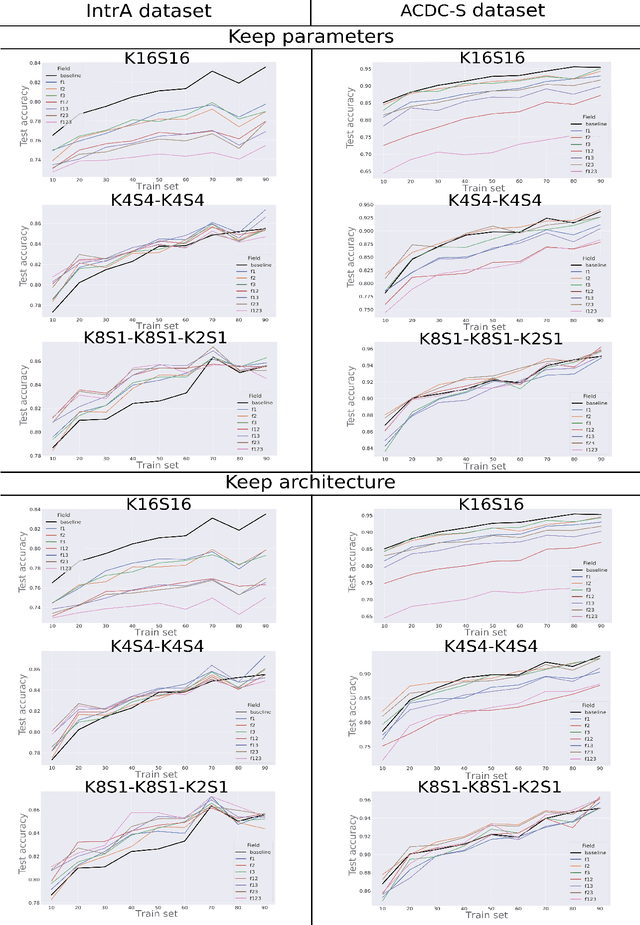
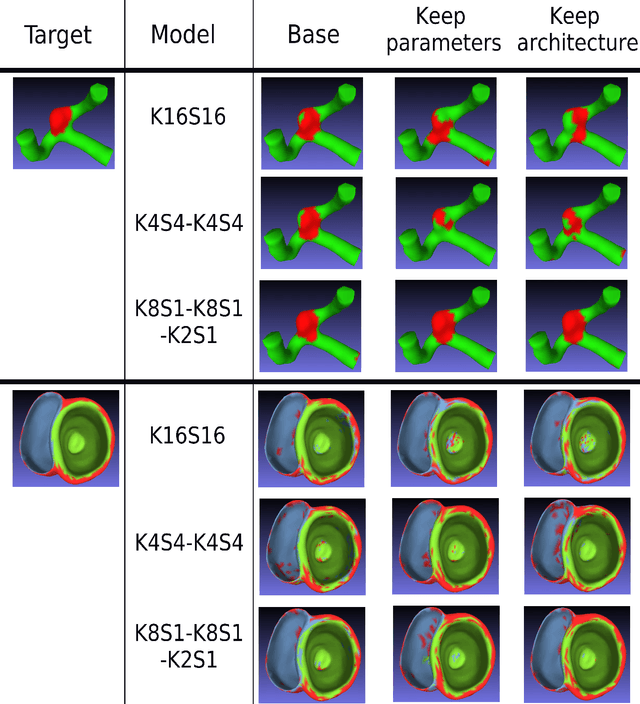
Abstract:3D mesh segmentation is an important task with many biomedical applications. The human body has bilateral symmetry and some variations in organ positions. It allows us to expect a positive effect of rotation and inversion invariant layers in convolutional neural networks that perform biomedical segmentations. In this study, we show the impact of weight symmetry in neural networks that perform 3D mesh segmentation. We analyze the problem of 3D mesh segmentation for pathological vessel structures (aneurysms) and conventional anatomical structures (endocardium and epicardium of ventricles). Local geometrical features are encoded as sampling from the signed distance function, and the neural network performs prediction for each mesh node. We show that weight symmetry gains from 1 to 3% of additional accuracy and allows decreasing the number of trainable parameters up to 8 times without suffering the performance loss if neural networks have at least three convolutional layers. This also works for very small training sets.
Compressor-Based Classification for Atrial Fibrillation Detection
Aug 25, 2023Abstract:Atrial fibrillation (AF) is one of the most common arrhythmias with challenging public health implications. Automatic detection of AF episodes is therefore one of the most important tasks in biomedical engineering. In this paper, we apply the recently introduced method of compressor-based text classification to the task of AF detection (binary classification between heart rhythms). We investigate the normalised compression distance applied to $\Delta$RR and RR-interval sequences, the configuration of the k-Nearest Neighbour classifier, and an optimal window length. We achieve good classification results (avg. sensitivity = 97.1%, avg. specificity = 91.7%, best sensitivity of 99.8%, best specificity of 97.6% with 5-fold cross-validation). Obtained performance is close to the best specialised AF detection algorithms. Our results suggest that gzip classification, originally proposed for texts, is suitable for biomedical data and continuous stochastic sequences in general.
Statistical model for describing heart rate variability in normal rhythm and atrial fibrillation
Jul 17, 2022
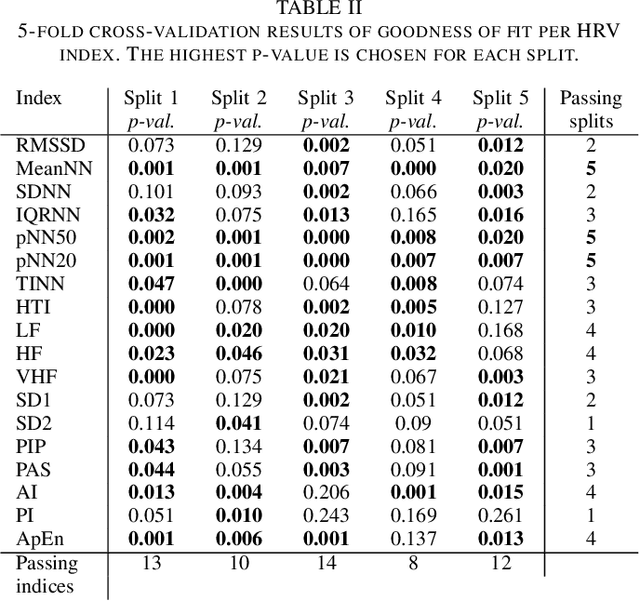
Abstract:Heart rate variability (HRV) indices describe properties of interbeat intervals in electrocardiogram (ECG). Usually HRV is measured exclusively in normal sinus rhythm (NSR) excluding any form of paroxysmal rhythm. Atrial fibrillation (AF) is the most widespread cardiac arrhythmia in human population. Usually such abnormal rhythm is not analyzed and assumed to be chaotic and unpredictable. Nonetheless, ranges of HRV indices differ between patients with AF, yet physiological characteristics which influence them are poorly understood. In this study, we propose a statistical model that describes relationship between HRV indices in NSR and AF. The model is based on Mahalanobis distance, the k-Nearest neighbour approach and multivariate normal distribution framework. Verification of the method was performed using 10 min intervals of NSR and AF that were extracted from long-term Holter ECGs. For validation we used Bhattacharyya distance and Kolmogorov-Smirnov 2-sample test in a k-fold procedure. The model is able to predict at least 7 HRV indices with high precision.
Natural language processing for clusterization of genes according to their functions
Jul 17, 2022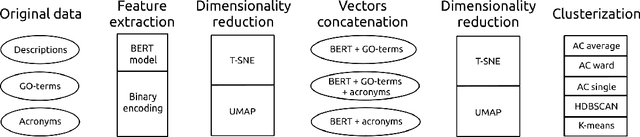
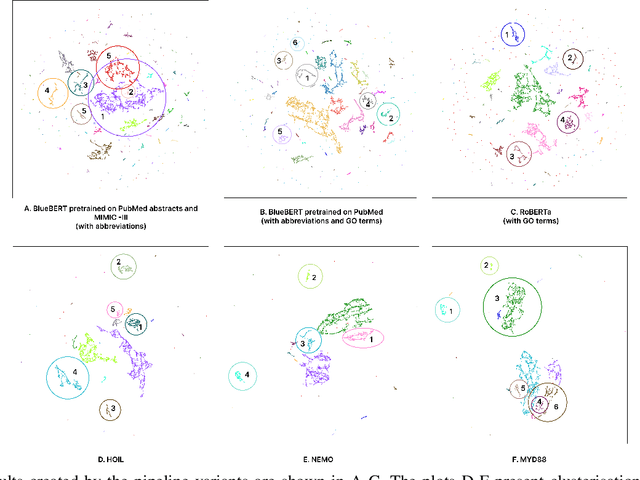
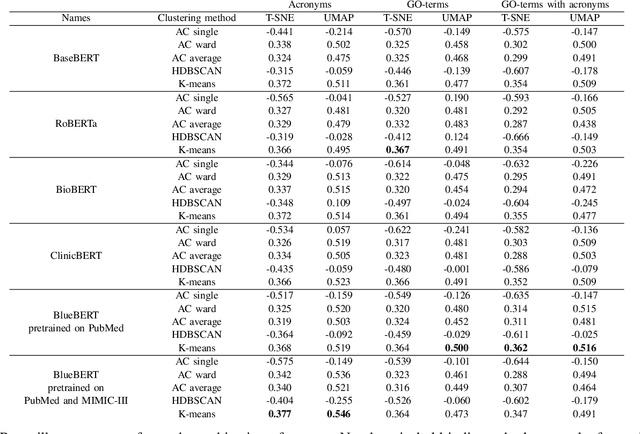
Abstract:There are hundreds of methods for analysis of data obtained in mRNA-sequencing. The most of them are focused on small number of genes. In this study, we propose an approach that reduces the analysis of several thousand genes to analysis of several clusters. The list of genes is enriched with information from open databases. Then, the descriptions are encoded as vectors using the pretrained language model (BERT) and some text processing approaches. The encoded gene function pass through the dimensionality reduction and clusterization. Aiming to find the most efficient pipeline, 180 cases of pipeline with different methods in the major pipeline steps were analyzed. The performance was evaluated with clusterization indexes and expert review of the results.
Anomaly Detection in Image Datasets Using Convolutional Neural Networks, Center Loss, and Mahalanobis Distance
Apr 13, 2021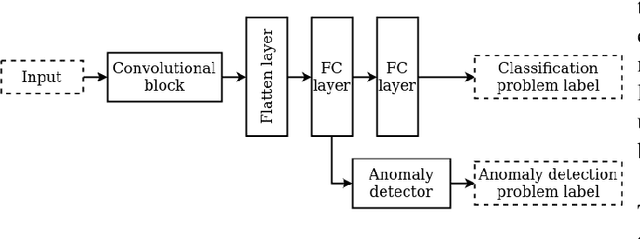
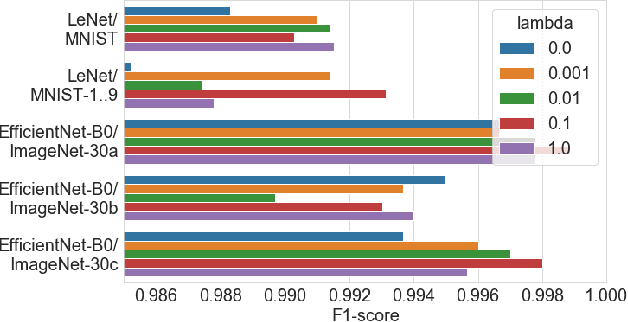
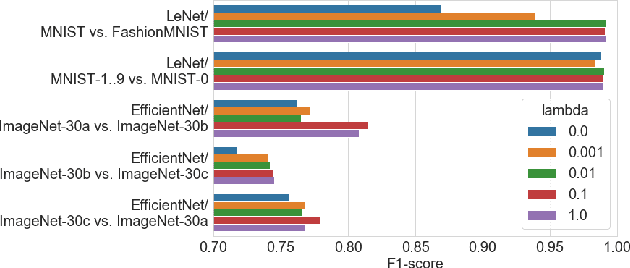
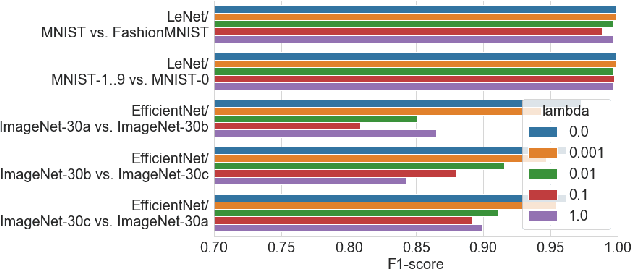
Abstract:User activities generate a significant number of poor-quality or irrelevant images and data vectors that cannot be processed in the main data processing pipeline or included in the training dataset. Such samples can be found with manual analysis by an expert or with anomalous detection algorithms. There are several formal definitions for the anomaly samples. For neural networks, the anomalous is usually defined as out-of-distribution samples. This work proposes methods for supervised and semi-supervised detection of out-of-distribution samples in image datasets. Our approach extends a typical neural network that solves the image classification problem. Thus, one neural network after extension can solve image classification and anomalous detection problems simultaneously. Proposed methods are based on the center loss and its effect on a deep feature distribution in a last hidden layer of the neural network. This paper provides an analysis of the proposed methods for the LeNet and EfficientNet-B0 on the MNIST and ImageNet-30 datasets.
Effects of lead position, cardiac rhythm variation and drug-induced QT prolongation on performance of machine learning methods for ECG processing
Dec 10, 2019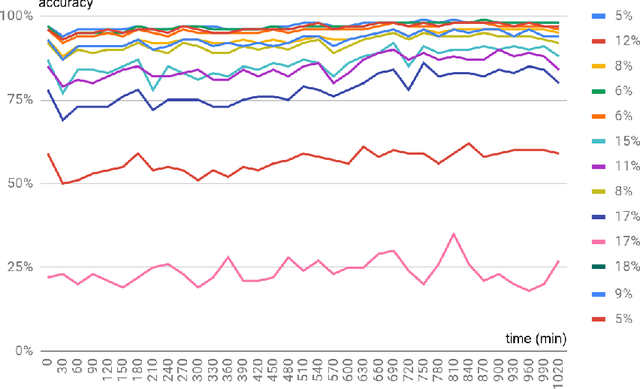
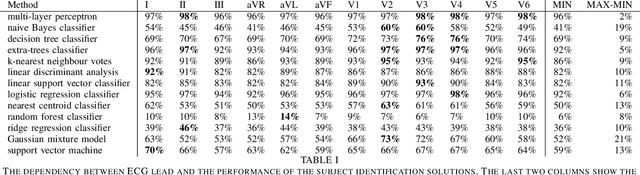
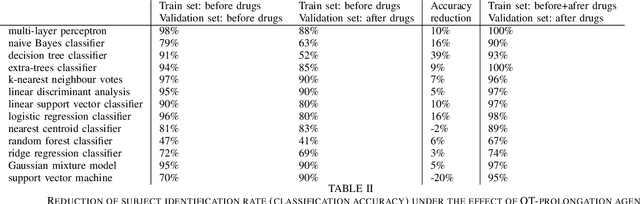
Abstract:Machine learning shows great performance in various problems of electrocardiography (ECG) signal analysis. However, collecting of any dataset for biomedical engineering is a very difficult task. Any datasets for ECG processing contains from 100 to 10,000 times fewer cases than datasets for image or text analysis. This issue is especially important because of physiological phenomena that can significantly change the morphology of heartbeats in ECG signals. In this preliminary study, we analyze the effects of lead choice from the standard ECG recordings, a variation of ECG during 24-hours, and the effects of QT-prolongation agents on the performance of machine learning methods for ECG processing. We choose the problem of subject identification for analysis, because this problem may be solved for almost any available dataset of ECG data. In a discussion, we compare our findings with observations from other works that use machine learning for ECG processing with different problem statements. Our results show the importance of training dataset enrichment with ECG signals that acquired in specific physiological conditions for obtaining good performance of ECG processing for real applications.
Phase mapping for cardiac unipolar electrograms with neural network instead of phase transformation
Nov 21, 2019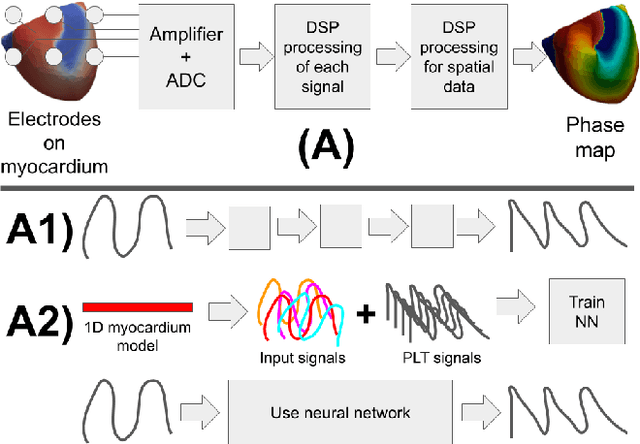


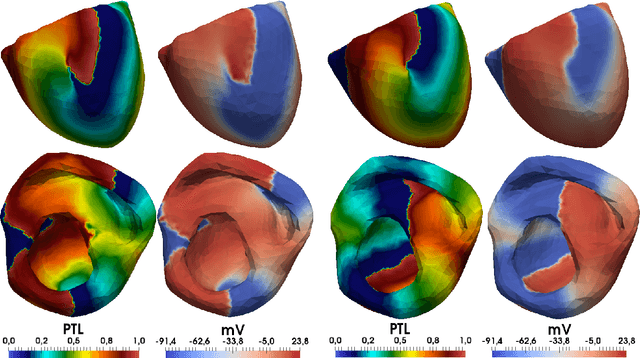
Abstract:Digital signal processing can be performed in two major ways. The first is a pipeline of signal transformations, and the second is machine learning approaches that require tagged data for training. This paper studies the third way. We generate a training dataset for a neural network in a series of numerical experiments and uses the trained neural network for processing of new signals. The current work focuses on phase mapping, which is an approach of cardiac unipolar electrogram processing, that helps to analyze complex non-stationary behavior of cardiac arrhythmias. Idealized models of 1D myocardial electrophysiology provide a training dataset. Then, the convolution neural network is trained to provide phase-like transformation for the phase mapping. The proposed approaches were validated against data from the detailed personalized model of human torso electrophysiology. The current paper includes a visualization of the phase map based on our approach and shows the robustness of the proposed approaches in the analysis of the complex non-stationary periodic activity of the excitable cardiac tissue.
Comparison of UNet, ENet, and BoxENet for Segmentation of Mast Cells in Scans of Histological Slices
Oct 15, 2019
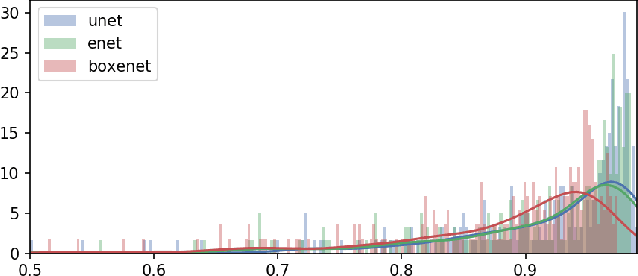
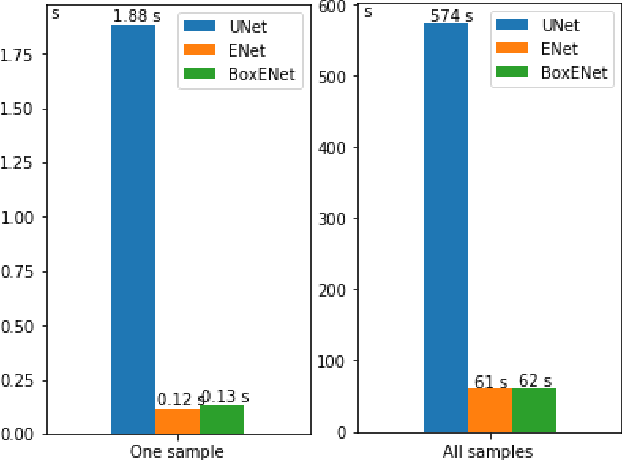

Abstract:Deep neural networks show high accuracy in theproblem of semantic and instance segmentation of biomedicaldata. However, this approach is computationally expensive. Thecomputational cost may be reduced with network simplificationafter training or choosing the proper architecture, which providessegmentation with less accuracy but does it much faster. In thepresent study, we analyzed the accuracy and performance ofUNet and ENet architectures for the problem of semantic imagesegmentation. In addition, we investigated the ENet architecture by replacing of some convolution layers with box-convolutionlayers. The analysis performed on the original dataset consisted of histology slices with mast cells. These cells provide a region forsegmentation with different types of borders, which vary fromclearly visible to ragged. ENet was less accurate than UNet byonly about 1-2%, but ENet performance was 8-15 times faster than UNet one.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge