Arlo Sheridan
Roadmap on Deep Learning for Microscopy
Mar 07, 2023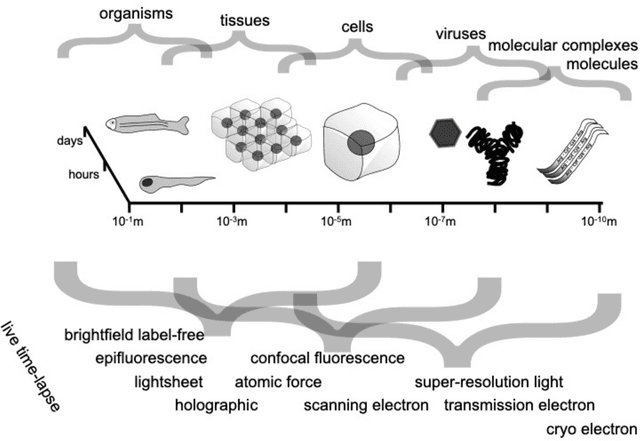
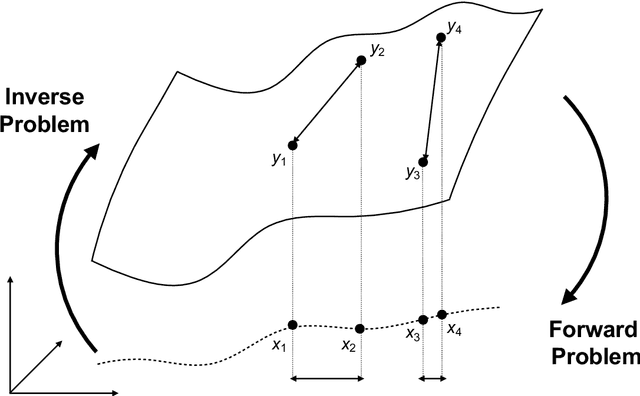
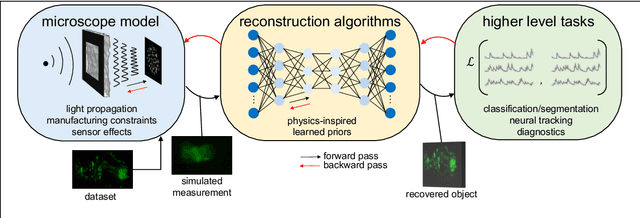
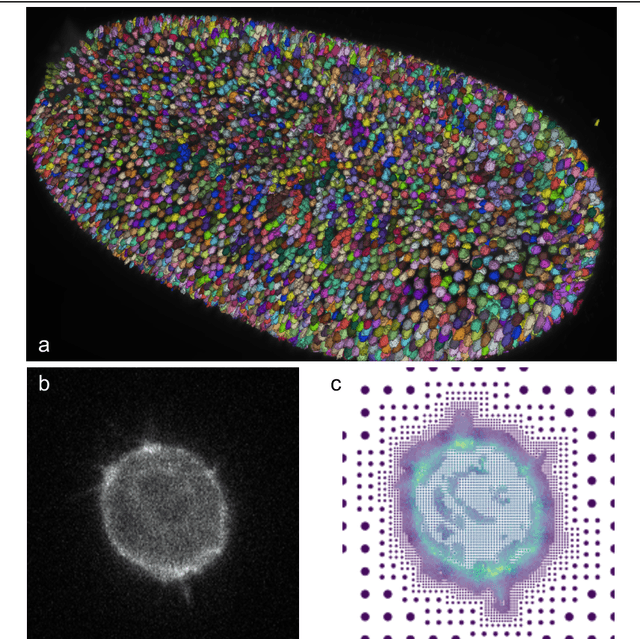
Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
A Deep Structured Learning Approach Towards Automating Connectome Reconstruction from 3D Electron Micrographs
Sep 24, 2017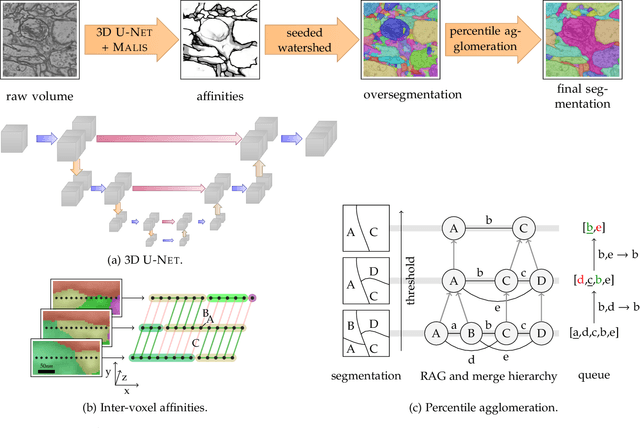

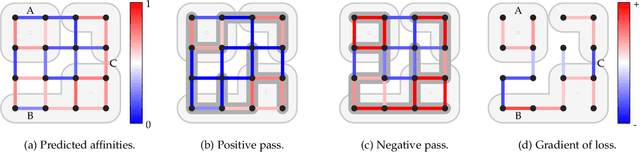

Abstract:We present a deep structured learning method for neuron segmentation from 3D electron microscopy (EM) which improves significantly upon the state of the art in terms of accuracy and scalability. Our method consists of a 3D U-Net classifier predicting affinity graphs on voxels, followed by iterative region agglomeration. We train the U-Net using a new structured loss based on MALIS that encourages topological correctness. Our extension consists of two parts: First, an $O(n\log(n))$ method to compute the loss gradient, improving over the originally proposed $O(n^2)$ algorithm. Second, we compute the gradient in two separate passes to avoid spurious contributions in early training stages. Our affinity predictions are accurate enough that simple agglomeration outperforms more involved methods used earlier on inferior predictions. We present results on three datasets (CREMI, FIB, and SegEM) of different imaging techniques and animals and achieve improvements over previous results of 27%, 15%, and 250%. Our findings suggest that a single 3D segmentation strategy can be applied to both isotropic and anisotropic EM data. The runtime of our method scales with $O(n)$ in the size of the volume and achieves a throughput of about 2.6 seconds per megavoxel, allowing processing of very large datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge