Arghya Bhowmik
Global Plane Waves From Local Gaussians: Periodic Charge Densities in a Blink
Jan 27, 2026Abstract:We introduce ELECTRAFI, a fast, end-to-end differentiable model for predicting periodic charge densities in crystalline materials. ELECTRAFI constructs anisotropic Gaussians in real space and exploits their closed-form Fourier transforms to analytically evaluate plane-wave coefficients via the Poisson summation formula. This formulation delegates non-local and periodic behavior to analytic transforms, enabling reconstruction of the full periodic charge density with a single inverse FFT. By avoiding explicit real-space grid probing, periodic image summation, and spherical harmonic expansions, ELECTRAFI matches or exceeds state-of-the-art accuracy across periodic benchmarks while being up to $633 \times$ faster than the strongest competing method, reconstructing crystal charge densities in a fraction of a second. When used to initialize DFT calculations, ELECTRAFI reduces total DFT compute cost by up to ~20%, whereas slower charge density models negate savings due to high inference times. Our results show that accuracy and inference cost jointly determine end-to-end DFT speedups, and motivate our focus on efficiency.
Reinforcement Learning for Chemical Ordering in Alloy Nanoparticles
Nov 15, 2025



Abstract:We approach the search for optimal element ordering in bimetallic alloy nanoparticles (NPs) as a reinforcement learning (RL) problem, and have built an RL agent that learns to perform such global optimisation using the geometric graph representation of the NPs. To demonstrate the effectiveness, we train an RL agent to perform composition-conserving atomic swap actions on the icosahedral nanoparticle structure. Trained once on randomised $Ag_{X}Au_{309-X}$ compositions and orderings, the agent discovers previously established ground state structure. We show that this optimization is robust to differently ordered initialisations of the same NP compositions. We also demonstrate that a trained policy can extrapolate effectively to NPs of unseen size. However, the efficacy is limited when multiple alloying elements are involved. Our results demonstrate that RL with pre-trained equivariant graph encodings can navigate combinatorial ordering spaces at the nanoparticle scale, and offer a transferable optimisation strategy with the potential to generalise across composition and reduce repeated individual search cost.
Acoustic Classification of Maritime Vessels using Learnable Filterbanks
May 29, 2025Abstract:Reliably monitoring and recognizing maritime vessels based on acoustic signatures is complicated by the variability of different recording scenarios. A robust classification framework must be able to generalize across diverse acoustic environments and variable source-sensor distances. To this end, we present a deep learning model with robust performance across different recording scenarios. Using a trainable spectral front-end and temporal feature encoder to learn a Gabor filterbank, the model can dynamically emphasize different frequency components. Trained on the VTUAD hydrophone recordings from the Strait of Georgia, our model, CATFISH, achieves a state-of-the-art 96.63 % percent test accuracy across varying source-sensor distances, surpassing the previous benchmark by over 12 percentage points. We present the model, justify our architectural choices, analyze the learned Gabor filters, and perform ablation studies on sensor data fusion and attention-based pooling.
ELECTRA: A Symmetry-breaking Cartesian Network for Charge Density Prediction with Floating Orbitals
Mar 11, 2025Abstract:We present the Electronic Tensor Reconstruction Algorithm (ELECTRA) - an equivariant model for predicting electronic charge densities using "floating" orbitals. Floating orbitals are a long-standing idea in the quantum chemistry community that promises more compact and accurate representations by placing orbitals freely in space, as opposed to centering all orbitals at the position of atoms. Finding ideal placements of these orbitals requires extensive domain knowledge though, which thus far has prevented widespread adoption. We solve this in a data-driven manner by training a Cartesian tensor network to predict orbital positions along with orbital coefficients. This is made possible through a symmetry-breaking mechanism that is used to learn position displacements with lower symmetry than the input molecule while preserving the rotation equivariance of the charge density itself. Inspired by recent successes of Gaussian Splatting in representing densities in space, we are using Gaussians as our orbitals and predict their weights and covariance matrices. Our method achieves a state-of-the-art balance between computational efficiency and predictive accuracy on established benchmarks.
Transition1x -- a Dataset for Building Generalizable Reactive Machine Learning Potentials
Jul 25, 2022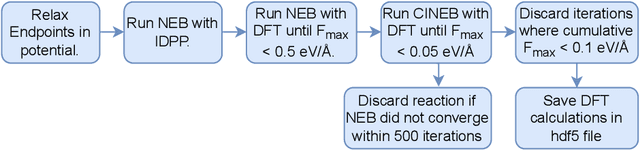
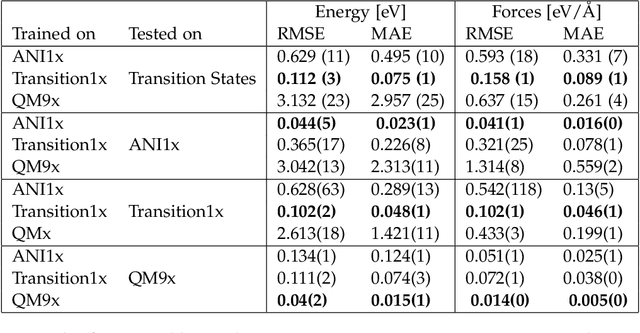
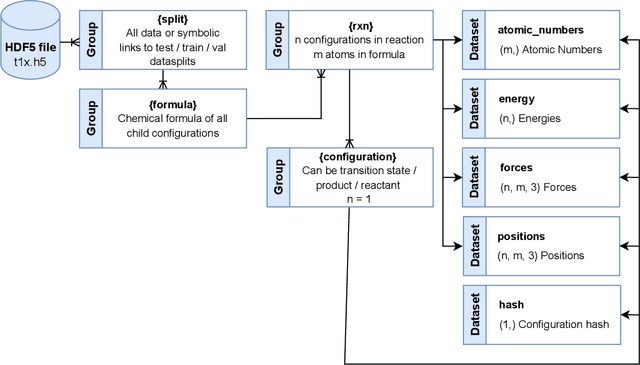
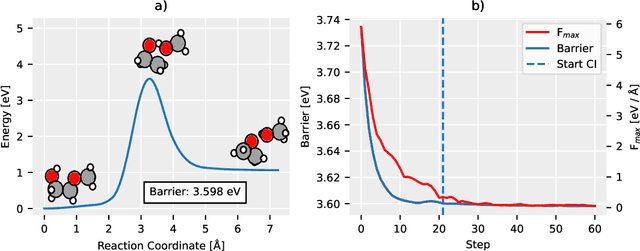
Abstract:Machine Learning (ML) models have, in contrast to their usefulness in molecular dynamics studies, had limited success as surrogate potentials for reaction barrier search. It is due to the scarcity of training data in relevant transition state regions of chemical space. Currently, available datasets for training ML models on small molecular systems almost exclusively contain configurations at or near equilibrium. In this work, we present the dataset Transition1x containing 9.6 million Density Functional Theory (DFT) calculations of forces and energies of molecular configurations on and around reaction pathways at the wB97x/6-31G(d) level of theory. The data was generated by running Nudged Elastic Band (NEB) calculations with DFT on 10k reactions while saving intermediate calculations. We train state-of-the-art equivariant graph message-passing neural network models on Transition1x and cross-validate on the popular ANI1x and QM9 datasets. We show that ML models cannot learn features in transition-state regions solely by training on hitherto popular benchmark datasets. Transition1x is a new challenging benchmark that will provide an important step towards developing next-generation ML force fields that also work far away from equilibrium configurations and reactive systems.
NeuralNEB -- Neural Networks can find Reaction Paths Fast
Jul 21, 2022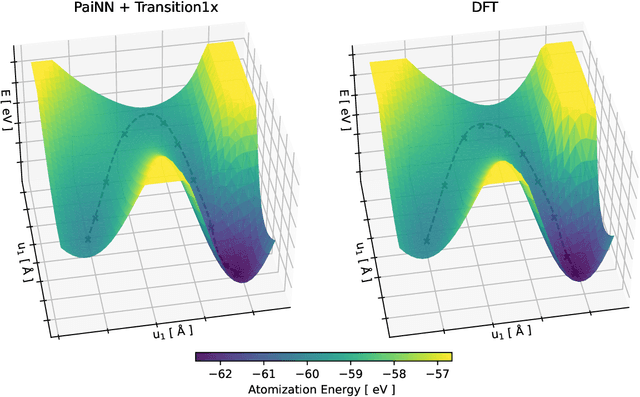
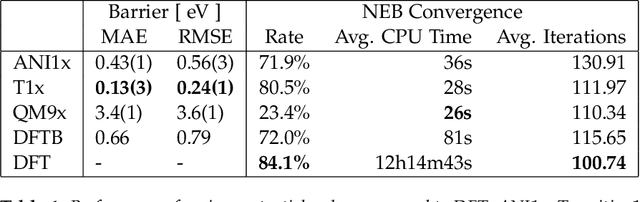
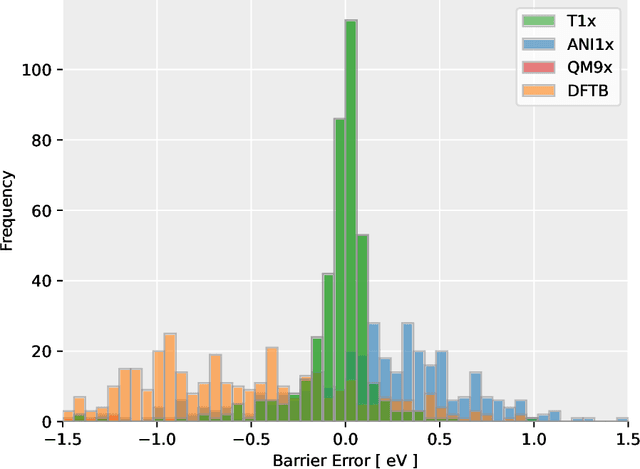
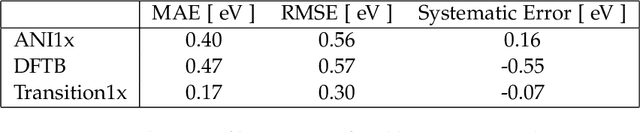
Abstract:Quantum mechanical methods like Density Functional Theory (DFT) are used with great success alongside efficient search algorithms for studying kinetics of reactive systems. However, DFT is prohibitively expensive for large scale exploration. Machine Learning (ML) models have turned out to be excellent emulators of small molecule DFT calculations and could possibly replace DFT in such tasks. For kinetics, success relies primarily on the models capability to accurately predict the Potential Energy Surface (PES) around transition-states and Minimal Energy Paths (MEPs). Previously this has not been possible due to scarcity of relevant data in the literature. In this paper we train state of the art equivariant Graph Neural Network (GNN)-based models on around 10.000 elementary reactions from the Transition1x dataset. We apply the models as potentials for the Nudged Elastic Band (NEB) algorithm and achieve a Mean Average Error (MAE) of 0.13+/-0.03 eV on barrier energies on unseen reactions. We compare the results against equivalent models trained on QM9 and ANI1x. We also compare with and outperform Density Functional based Tight Binding (DFTB) on both accuracy and computational resource. The implication is that ML models, given relevant data, are now at a level where they can be applied for downstream tasks in quantum chemistry transcending prediction of simple molecular features.
Graph neural networks for fast electron density estimation of molecules, liquids, and solids
Dec 01, 2021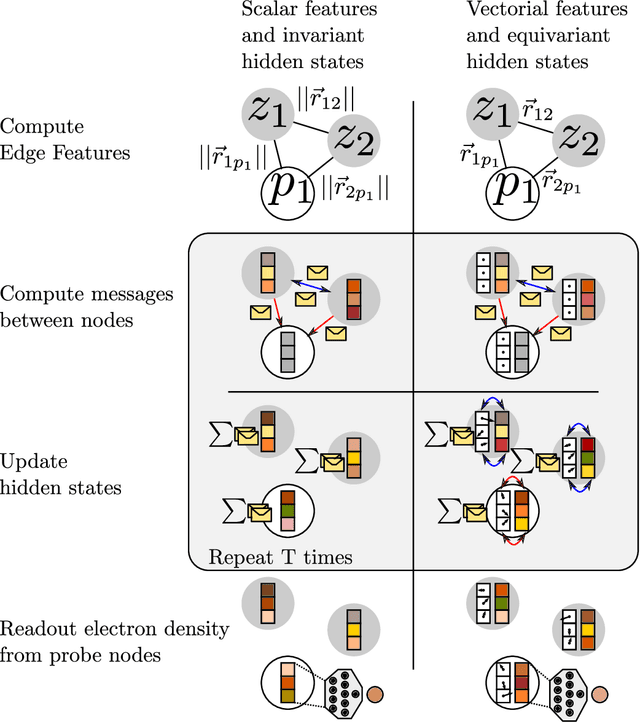
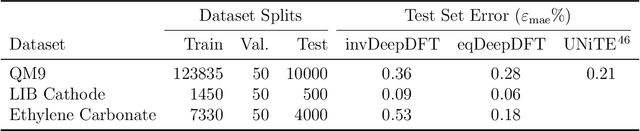
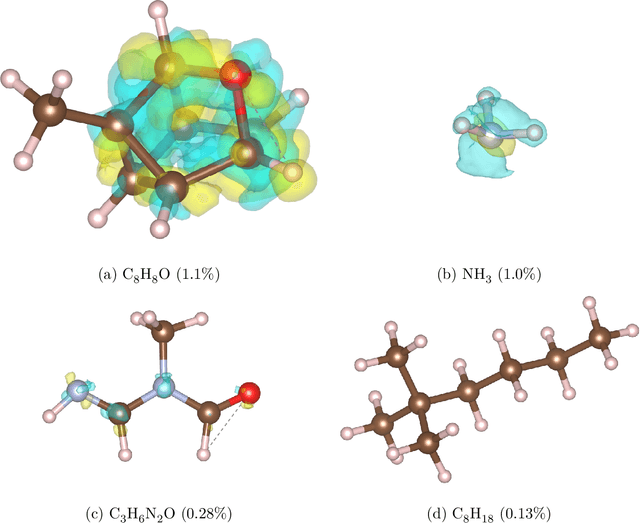
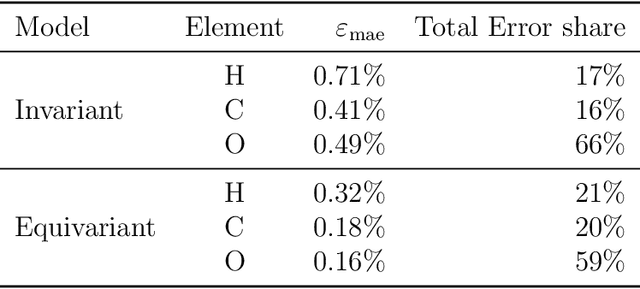
Abstract:Electron density $\rho(\vec{r})$ is the fundamental variable in the calculation of ground state energy with density functional theory (DFT). Beyond total energy, features in $\rho(\vec{r})$ distribution and modifications in $\rho(\vec{r})$ are often used to capture critical physicochemical phenomena in functional materials and molecules at the electronic scale. Methods providing access to $\rho(\vec{r})$ of complex disordered systems with little computational cost can be a game changer in the expedited exploration of materials phase space towards the inverse design of new materials with better functionalities. We present a machine learning framework for the prediction of $\rho(\vec{r})$. The model is based on equivariant graph neural networks and the electron density is predicted at special query point vertices that are part of the message passing graph, but only receive messages. The model is tested across multiple data sets of molecules (QM9), liquid ethylene carbonate electrolyte (EC) and LixNiyMnzCo(1-y-z)O2 lithium ion battery cathodes (NMC). For QM9 molecules, the accuracy of the proposed model exceeds typical variability in $\rho(\vec{r})$ obtained from DFT done with different exchange-correlation functional and show beyond the state of the art accuracy. The accuracy is even better for the mixed oxide (NMC) and electrolyte (EC) datasets. The linear scaling model's capacity to probe thousands of points simultaneously permits calculation of $\rho(\vec{r})$ for large complex systems many orders of magnitude faster than DFT allowing screening of disordered functional materials.
Calibrated Uncertainty for Molecular Property Prediction using Ensembles of Message Passing Neural Networks
Jul 13, 2021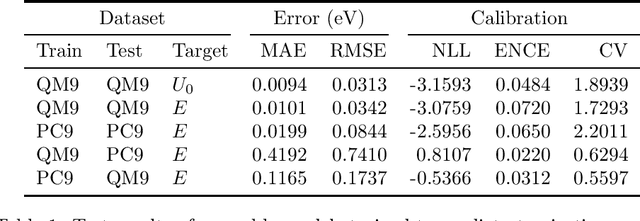

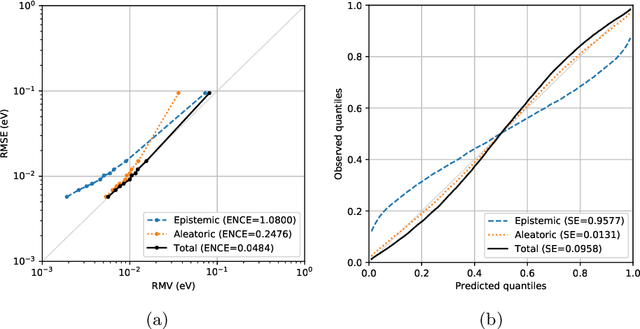
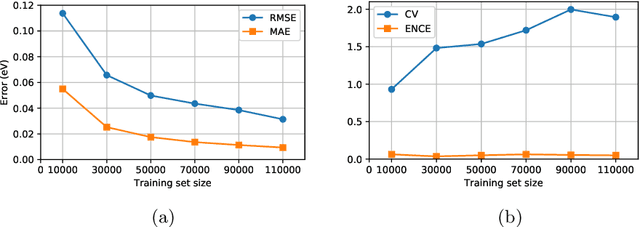
Abstract:Data-driven methods based on machine learning have the potential to accelerate analysis of atomic structures. However, machine learning models can produce overconfident predictions and it is therefore crucial to detect and handle uncertainty carefully. Here, we extend a message passing neural network designed specifically for predicting properties of molecules and materials with a calibrated probabilistic predictive distribution. The method presented in this paper differs from the previous work by considering both aleatoric and epistemic uncertainty in a unified framework, and by re-calibrating the predictive distribution on unseen data. Through computer experiments, we show that our approach results in accurate models for predicting molecular formation energies with calibrated uncertainty in and out of the training data distribution on two public molecular benchmark datasets, QM9 and PC9. The proposed method provides a general framework for training and evaluating neural network ensemble models that are able to produce accurate predictions of properties of molecules with calibrated uncertainty.
DeepDFT: Neural Message Passing Network for Accurate Charge Density Prediction
Nov 04, 2020
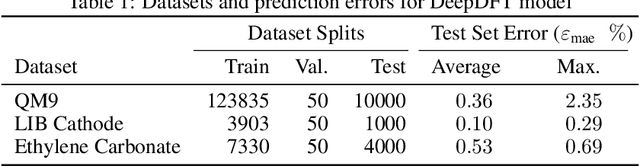
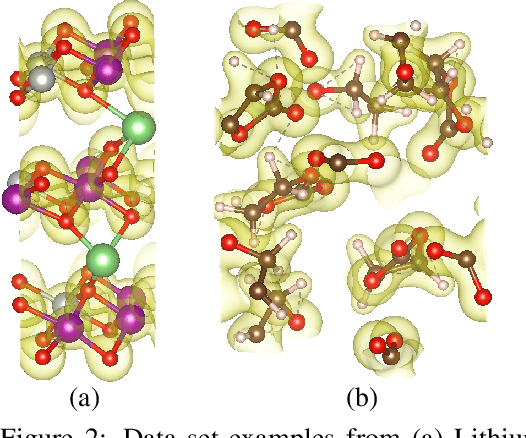
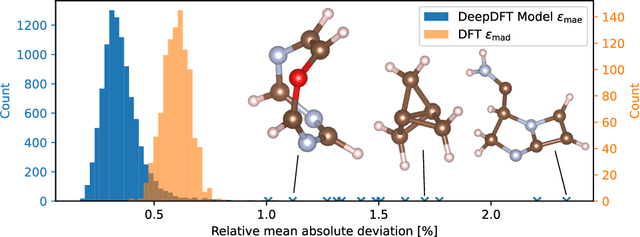
Abstract:We introduce DeepDFT, a deep learning model for predicting the electronic charge density around atoms, the fundamental variable in electronic structure simulations from which all ground state properties can be calculated. The model is formulated as neural message passing on a graph, consisting of interacting atom vertices and special query point vertices for which the charge density is predicted. The accuracy and scalability of the model are demonstrated for molecules, solids and liquids. The trained model achieves lower average prediction errors than the observed variations in charge density obtained from density functional theory simulations using different exchange correlation functionals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge