Anastasia Razdaibiedina
The Llama 4 Herd: Architecture, Training, Evaluation, and Deployment Notes
Jan 15, 2026Abstract:This document consolidates publicly reported technical details about Metas Llama 4 model family. It summarizes (i) released variants (Scout and Maverick) and the broader herd context including the previewed Behemoth teacher model, (ii) architectural characteristics beyond a high-level MoE description covering routed/shared-expert structure, early-fusion multimodality, and long-context design elements reported for Scout (iRoPE and length generalization strategies), (iii) training disclosures spanning pre-training, mid-training for long-context extension, and post-training methodology (lightweight SFT, online RL, and lightweight DPO) as described in release materials, (iv) developer-reported benchmark results for both base and instruction-tuned checkpoints, and (v) practical deployment constraints observed across major serving environments, including provider-specific context limits and quantization packaging. The manuscript also summarizes licensing obligations relevant to redistribution and derivative naming, and reviews publicly described safeguards and evaluation practices. The goal is to provide a compact technical reference for researchers and practitioners who need precise, source-backed facts about Llama 4.
Orca 2: Teaching Small Language Models How to Reason
Nov 21, 2023



Abstract:Orca 1 learns from rich signals, such as explanation traces, allowing it to outperform conventional instruction-tuned models on benchmarks like BigBench Hard and AGIEval. In Orca 2, we continue exploring how improved training signals can enhance smaller LMs' reasoning abilities. Research on training small LMs has often relied on imitation learning to replicate the output of more capable models. We contend that excessive emphasis on imitation may restrict the potential of smaller models. We seek to teach small LMs to employ different solution strategies for different tasks, potentially different from the one used by the larger model. For example, while larger models might provide a direct answer to a complex task, smaller models may not have the same capacity. In Orca 2, we teach the model various reasoning techniques (step-by-step, recall then generate, recall-reason-generate, direct answer, etc.). More crucially, we aim to help the model learn to determine the most effective solution strategy for each task. We evaluate Orca 2 using a comprehensive set of 15 diverse benchmarks (corresponding to approximately 100 tasks and over 36,000 unique prompts). Orca 2 significantly surpasses models of similar size and attains performance levels similar or better to those of models 5-10x larger, as assessed on complex tasks that test advanced reasoning abilities in zero-shot settings. make Orca 2 weights publicly available at aka.ms/orca-lm to support research on the development, evaluation, and alignment of smaller LMs
MIReAD: Simple Method for Learning High-quality Representations from Scientific Documents
May 07, 2023Abstract:Learning semantically meaningful representations from scientific documents can facilitate academic literature search and improve performance of recommendation systems. Pre-trained language models have been shown to learn rich textual representations, yet they cannot provide powerful document-level representations for scientific articles. We propose MIReAD, a simple method that learns high-quality representations of scientific papers by fine-tuning transformer model to predict the target journal class based on the abstract. We train MIReAD on more than 500,000 PubMed and arXiv abstracts across over 2,000 journal classes. We show that MIReAD produces representations that can be used for similar papers retrieval, topic categorization and literature search. Our proposed approach outperforms six existing models for representation learning on scientific documents across four evaluation standards.
Residual Prompt Tuning: Improving Prompt Tuning with Residual Reparameterization
May 06, 2023



Abstract:Prompt tuning is one of the successful approaches for parameter-efficient tuning of pre-trained language models. Despite being arguably the most parameter-efficient (tuned soft prompts constitute <0.1% of total parameters), it typically performs worse than other efficient tuning methods and is quite sensitive to hyper-parameters. In this work, we introduce Residual Prompt Tuning - a simple and efficient method that significantly improves the performance and stability of prompt tuning. We propose to reparameterize soft prompt embeddings using a shallow network with a residual connection. Our experiments show that Residual Prompt Tuning significantly outperforms prompt tuning on SuperGLUE benchmark. Notably, our method reaches +7 points improvement over prompt tuning with T5-Base and allows to reduce the prompt length by 10x without hurting performance. In addition, we show that our approach is robust to the choice of learning rate and prompt initialization, and is effective in few-shot settings.
Progressive Prompts: Continual Learning for Language Models
Jan 29, 2023



Abstract:We introduce Progressive Prompts - a simple and efficient approach for continual learning in language models. Our method allows forward transfer and resists catastrophic forgetting, without relying on data replay or a large number of task-specific parameters. Progressive Prompts learns a new soft prompt for each task and sequentially concatenates it with the previously learned prompts, while keeping the base model frozen. Experiments on standard continual learning benchmarks show that our approach outperforms state-of-the-art methods, with an improvement >20% in average test accuracy over the previous best-preforming method on T5 model. We also explore a more challenging continual learning setup with longer sequences of tasks and show that Progressive Prompts significantly outperforms prior methods.
Learning multi-scale functional representations of proteins from single-cell microscopy data
May 24, 2022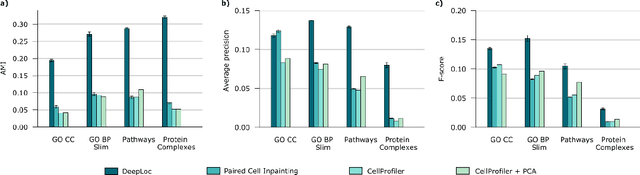
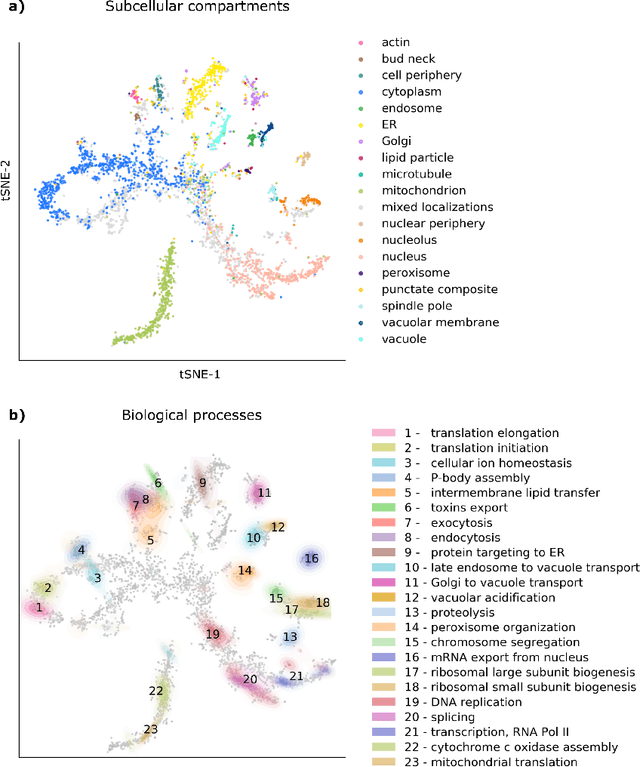
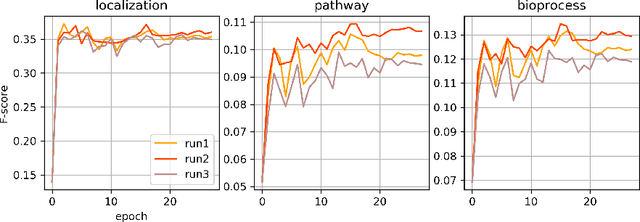
Abstract:Protein function is inherently linked to its localization within the cell, and fluorescent microscopy data is an indispensable resource for learning representations of proteins. Despite major developments in molecular representation learning, extracting functional information from biological images remains a non-trivial computational task. Current state-of-the-art approaches use autoencoder models to learn high-quality features by reconstructing images. However, such methods are prone to capturing noise and imaging artifacts. In this work, we revisit deep learning models used for classifying major subcellular localizations, and evaluate representations extracted from their final layers. We show that simple convolutional networks trained on localization classification can learn protein representations that encapsulate diverse functional information, and significantly outperform autoencoder-based models. We also propose a robust evaluation strategy to assess quality of protein representations across different scales of biological function.
Improving language models fine-tuning with representation consistency targets
May 23, 2022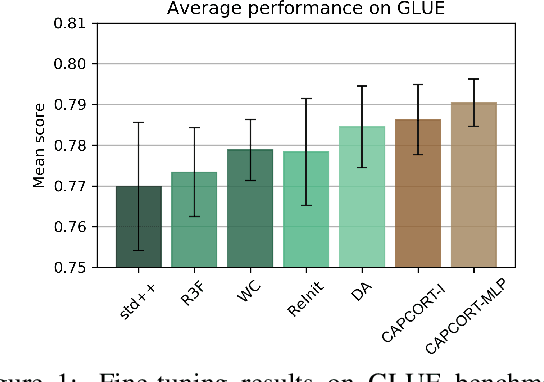

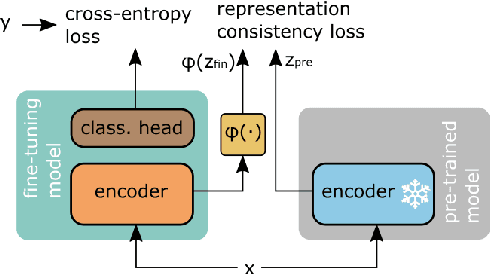

Abstract:Fine-tuning contextualized representations learned by pre-trained language models has become a standard practice in the NLP field. However, pre-trained representations are prone to degradation (also known as representation collapse) during fine-tuning, which leads to instability, suboptimal performance, and weak generalization. In this paper, we propose a novel fine-tuning method that avoids representation collapse during fine-tuning by discouraging undesirable changes in the representations. We show that our approach matches or exceeds the performance of the existing regularization-based fine-tuning methods across 13 language understanding tasks (GLUE benchmark and six additional datasets). We also demonstrate its effectiveness in low-data settings and robustness to label perturbation. Furthermore, we extend previous studies of representation collapse and propose several metrics to quantify it. Using these metrics and previously proposed experiments, we show that our approach obtains significant improvements in retaining the expressive power of representations.
Multi-defect microscopy image restoration under limited data conditions
Oct 31, 2019



Abstract:Deep learning methods are becoming widely used for restoration of defects associated with fluorescence microscopy imaging. One of the major challenges in application of such methods is the availability of training data. In this work, we pro-pose a unified method for reconstruction of multi-defect fluorescence microscopy images when training data is limited. Our approach consists of two steps: first, we perform data augmentation using Generative Adversarial Network (GAN) with conditional instance normalization (CIN); second, we train a conditional GAN(cGAN) on paired ground-truth and defected images to perform restoration. The experiments on three common types of imaging defects with different amounts of training data, show that the proposed method gives comparable results or outperforms CARE, deblurGAN and CycleGAN in restored image quality when limited data is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge