Alzheimer's Disease Neuroimaging Initiative
IGCN: Integrative Graph Convolutional Networks for Multi-modal Data
Feb 04, 2024Abstract:Recent advances in Graph Neural Networks (GNN) have led to a considerable growth in graph data modeling for multi-modal data which contains various types of nodes and edges. Although some integrative prediction solutions have been developed recently for network-structured data, these methods have some restrictions. For a node classification task involving multi-modal data, certain data modalities may perform better when predicting one class, while others might excel in predicting a different class. Thus, to obtain a better learning representation, advanced computational methodologies are required for the integrative analysis of multi-modal data. Moreover, existing integrative tools lack a comprehensive and cohesive understanding of the rationale behind their specific predictions, making them unsuitable for enhancing model interpretability. Addressing these restrictions, we introduce a novel integrative neural network approach for multi-modal data networks, named Integrative Graph Convolutional Networks (IGCN). IGCN learns node embeddings from multiple topologies and fuses the multiple node embeddings into a weighted form by assigning attention coefficients to the node embeddings. Our proposed attention mechanism helps identify which types of data receive more emphasis for each sample to predict a certain class. Therefore, IGCN has the potential to unravel previously unknown characteristics within different node classification tasks. We benchmarked IGCN on several datasets from different domains, including a multi-omics dataset to predict cancer subtypes and a multi-modal clinical dataset to predict the progression of Alzheimer's disease. Experimental results show that IGCN outperforms or is on par with the state-of-the-art and baseline methods.
Thalamic nuclei segmentation from T$_1$-weighted MRI: unifying and benchmarking state-of-the-art methods with young and old cohorts
Sep 26, 2023Abstract:The thalamus and its constituent nuclei are critical for a broad range of cognitive and sensorimotor processes, and implicated in many neurological and neurodegenerative conditions. However, the functional involvement and specificity of thalamic nuclei in human neuroimaging is underappreciated and not well studied due, in part, to technical challenges of accurately identifying and segmenting nuclei. This challenge is further exacerbated by a lack of common nomenclature for comparing segmentation methods. Here, we use data from healthy young (Human Connectome Project, 100 subjects) and older healthy adults, plus those with minor cognitive impairment and Alzheimer$'$s disease (Alzheimer$'$s Disease Neuroimaging Initiative, 540 subjects), to benchmark four state of the art thalamic segmentation methods for T1 MRI (FreeSurfer, HIPS-THOMAS, SCS-CNN, and T1-THOMAS) under a single segmentation framework. Segmentations were compared using overlap and dissimilarity metrics to the Morel stereotaxic atlas. We also quantified each method$'$s estimation of thalamic nuclear degeneration across Alzheimer$'$s disease progression, and how accurately early and late mild cognitive impairment, and Alzheimers disease could be distinguished from healthy controls. We show that HIPS-THOMAS produced the most effective segmentations of individual thalamic nuclei and was also most accurate in discriminating healthy controls from those with mild cognitive impairment and Alzheimer$'$s disease using individual nucleus volumes. This work is the first to systematically compare the efficacy of anatomical thalamic segmentation approaches under a unified nomenclature. We also provide recommendations of which segmentation method to use for studying the functional relevance of specific thalamic nuclei, based on their overlap and dissimilarity with the Morel atlas.
Revisiting convolutional neural network on graphs with polynomial approximations of Laplace-Beltrami spectral filtering
Oct 26, 2020



Abstract:This paper revisits spectral graph convolutional neural networks (graph-CNNs) given in Defferrard (2016) and develops the Laplace-Beltrami CNN (LB-CNN) by replacing the graph Laplacian with the LB operator. We then define spectral filters via the LB operator on a graph. We explore the feasibility of Chebyshev, Laguerre, and Hermite polynomials to approximate LB-based spectral filters and define an update of the LB operator for pooling in the LBCNN. We employ the brain image data from Alzheimer's Disease Neuroimaging Initiative (ADNI) and demonstrate the use of the proposed LB-CNN. Based on the cortical thickness of the ADNI dataset, we showed that the LB-CNN didn't improve classification accuracy compared to the spectral graph-CNN. The three polynomials had a similar computational cost and showed comparable classification accuracy in the LB-CNN or spectral graph-CNN. Our findings suggest that even though the shapes of the three polynomials are different, deep learning architecture allows us to learn spectral filters such that the classification performance is not dependent on the type of the polynomials or the operators (graph Laplacian and LB operator).
Fast Mesh Data Augmentation via Chebyshev Polynomial of Spectral filtering
Oct 06, 2020



Abstract:Deep neural networks have recently been recognized as one of the powerful learning techniques in computer vision and medical image analysis. Trained deep neural networks need to be generalizable to new data that was not seen before. In practice, there is often insufficient training data available and augmentation is used to expand the dataset. Even though graph convolutional neural network (graph-CNN) has been widely used in deep learning, there is a lack of augmentation methods to generate data on graphs or surfaces. This study proposes two unbiased augmentation methods, Laplace-Beltrami eigenfunction Data Augmentation (LB-eigDA) and Chebyshev polynomial Data Augmentation (C-pDA), to generate new data on surfaces, whose mean is the same as that of real data. LB-eigDA augments data via the resampling of the LB coefficients. In parallel with LB-eigDA, we introduce a fast augmentation approach, C-pDA, that employs a polynomial approximation of LB spectral filters on surfaces. We design LB spectral bandpass filters by Chebyshev polynomial approximation and resample signals filtered via these filters to generate new data on surfaces. We first validate LB-eigDA and C-pDA via simulated data and demonstrate their use for improving classification accuracy. We then employ the brain images of Alzheimer's Disease Neuroimaging Initiative (ADNI) and extract cortical thickness that is represented on the cortical surface to illustrate the use of the two augmentation methods. We demonstrate that augmented cortical thickness has a similar pattern to real data. Second, we show that C-pDA is much faster than LB-eigDA. Last, we show that C-pDA can improve the AD classification accuracy of graph-CNN.
GANDALF: Generative Adversarial Networks with Discriminator-Adaptive Loss Fine-tuning for Alzheimer's Disease Diagnosis from MRI
Aug 10, 2020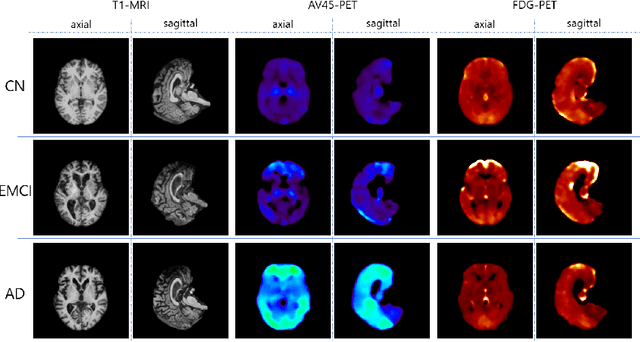
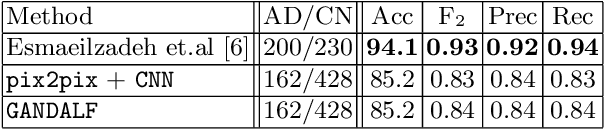
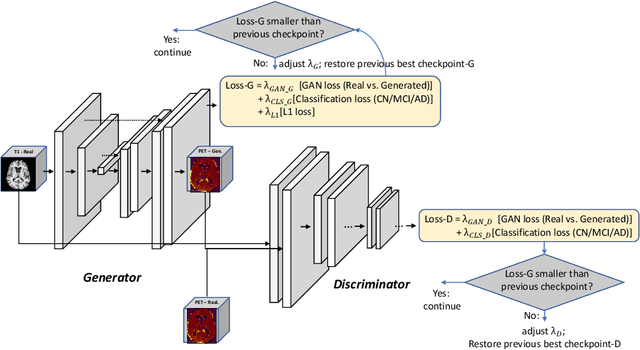
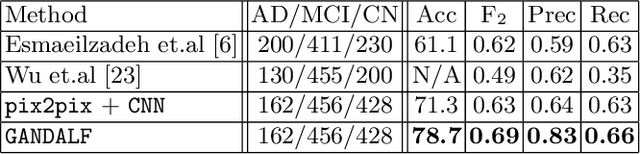
Abstract:Positron Emission Tomography (PET) is now regarded as the gold standard for the diagnosis of Alzheimer's Disease (AD). However, PET imaging can be prohibitive in terms of cost and planning, and is also among the imaging techniques with the highest dosage of radiation. Magnetic Resonance Imaging (MRI), in contrast, is more widely available and provides more flexibility when setting the desired image resolution. Unfortunately, the diagnosis of AD using MRI is difficult due to the very subtle physiological differences between healthy and AD subjects visible on MRI. As a result, many attempts have been made to synthesize PET images from MR images using generative adversarial networks (GANs) in the interest of enabling the diagnosis of AD from MR. Existing work on PET synthesis from MRI has largely focused on Conditional GANs, where MR images are used to generate PET images and subsequently used for AD diagnosis. There is no end-to-end training goal. This paper proposes an alternative approach to the aforementioned, where AD diagnosis is incorporated in the GAN training objective to achieve the best AD classification performance. Different GAN lossesare fine-tuned based on the discriminator performance, and the overall training is stabilized. The proposed network architecture and training regime show state-of-the-art performance for three- and four- class AD classification tasks.
GANBERT: Generative Adversarial Networks with Bidirectional Encoder Representations from Transformers for MRI to PET synthesis
Aug 10, 2020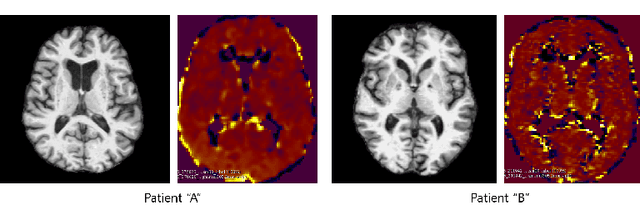
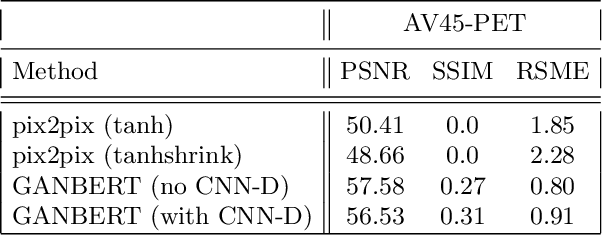
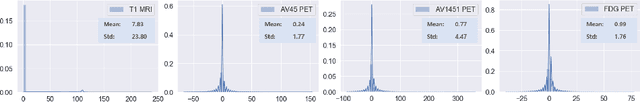
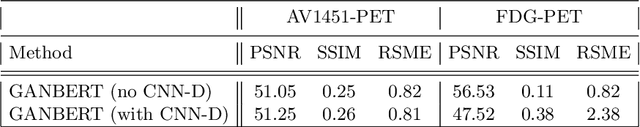
Abstract:Synthesizing medical images, such as PET, is a challenging task due to the fact that the intensity range is much wider and denser than those in photographs and digital renderings and are often heavily biased toward zero. Above all, intensity values in PET have absolute significance, and are used to compute parameters that are reproducible across the population. Yet, usually much manual adjustment has to be made in pre-/post- processing when synthesizing PET images, because its intensity ranges can vary a lot, e.g., between -100 to 1000 in floating point values. To overcome these challenges, we adopt the Bidirectional Encoder Representations from Transformers (BERT) algorithm that has had great success in natural language processing (NLP), where wide-range floating point intensity values are represented as integers ranging between 0 to 10000 that resemble a dictionary of natural language vocabularies. BERT is then trained to predict a proportion of masked values images, where its "next sentence prediction (NSP)" acts as GAN discriminator. Our proposed approach, is able to generate PET images from MRI images in wide intensity range, with no manual adjustments in pre-/post- processing. It is a method that can scale and ready to deploy.
An Optimized PatchMatch for Multi-scale and Multi-feature Label Fusion
Mar 17, 2019


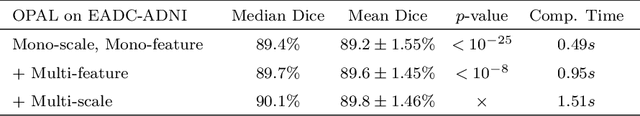
Abstract:Automatic segmentation methods are important tools for quantitative analysis of Magnetic Resonance Images (MRI). Recently, patch-based label fusion approaches have demonstrated state-of-the-art segmentation accuracy. In this paper, we introduce a new patch-based label fusion framework to perform segmentation of anatomical structures. The proposed approach uses an Optimized PAtchMatch Label fusion (OPAL) strategy that drastically reduces the computation time required for the search of similar patches. The reduced computation time of OPAL opens the way for new strategies and facilitates processing on large databases. In this paper, we investigate new perspectives offered by OPAL, by introducing a new multi-scale and multi-feature framework. During our validation on hippocampus segmentation we use two datasets: young adults in the ICBM cohort and elderly adults in the EADC-ADNI dataset. For both, OPAL is compared to state-of-the-art methods. Results show that OPAL obtained the highest median Dice coefficient (89.9% for ICBM and 90.1% for EADC-ADNI). Moreover, in both cases, OPAL produced a segmentation accuracy similar to inter-expert variability. On the EADC-ADNI dataset, we compare the hippocampal volumes obtained by manual and automatic segmentation. The volumes appear to be highly correlated that enables to perform more accurate separation of pathological populations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge