Manojkumar Saranathan
Department of Electrical and Computer Engineering, University of Arizona, Department of Medical Imaging, University of Arizona
Comprehensive segmentation of deep grey nuclei from structural MRI data
Mar 27, 2025



Abstract:Motivation: Lack of tools for comprehensive and complete segmentation of deep grey nuclei using a single software for reproducibility and repeatability Goal(s): A fast accurate and robust method for segmentation of deep grey nuclei (thalamic nuclei, basal ganglia, claustrum, red nucleus) from structural T1 MRI data at conventional field strengths Approach: We leverage the improved contrast of white-matter-nulled imaging by using the recently proposed Histogram-based Polynomial Synthesis (HIPS) to synthesize WMn-like images from standard T1 and then use a multi-atlas segmentation with joint label fusion to segment deep grey nuclei. Results: The method worked robustly on all field strengths (1.5/3/7) and Dice coefficients of 0.7 or more were achieved for all structures compared against manual segmentation ground truth. Impact: This method facilitates careful investigation of the role of deep grey nuclei by enabling the use of conventional T1 data from large public databases, which has not been possible, hitherto, due to lack of robust reproducible segmentation tools.
Thalamic nuclei segmentation from T$_1$-weighted MRI: unifying and benchmarking state-of-the-art methods with young and old cohorts
Sep 26, 2023Abstract:The thalamus and its constituent nuclei are critical for a broad range of cognitive and sensorimotor processes, and implicated in many neurological and neurodegenerative conditions. However, the functional involvement and specificity of thalamic nuclei in human neuroimaging is underappreciated and not well studied due, in part, to technical challenges of accurately identifying and segmenting nuclei. This challenge is further exacerbated by a lack of common nomenclature for comparing segmentation methods. Here, we use data from healthy young (Human Connectome Project, 100 subjects) and older healthy adults, plus those with minor cognitive impairment and Alzheimer$'$s disease (Alzheimer$'$s Disease Neuroimaging Initiative, 540 subjects), to benchmark four state of the art thalamic segmentation methods for T1 MRI (FreeSurfer, HIPS-THOMAS, SCS-CNN, and T1-THOMAS) under a single segmentation framework. Segmentations were compared using overlap and dissimilarity metrics to the Morel stereotaxic atlas. We also quantified each method$'$s estimation of thalamic nuclear degeneration across Alzheimer$'$s disease progression, and how accurately early and late mild cognitive impairment, and Alzheimers disease could be distinguished from healthy controls. We show that HIPS-THOMAS produced the most effective segmentations of individual thalamic nuclei and was also most accurate in discriminating healthy controls from those with mild cognitive impairment and Alzheimer$'$s disease using individual nucleus volumes. This work is the first to systematically compare the efficacy of anatomical thalamic segmentation approaches under a unified nomenclature. We also provide recommendations of which segmentation method to use for studying the functional relevance of specific thalamic nuclei, based on their overlap and dissimilarity with the Morel atlas.
Robust thalamic nuclei segmentation from T1-weighted MRI
Apr 14, 2023Abstract:Accurate segmentation of thalamic nuclei, crucial for understanding their role in healthy cognition and in pathologies, is challenging to achieve on standard T1-weighted (T1w) magnetic resonance imaging (MRI) due to poor image contrast. White-matter-nulled (WMn) MRI sequences improve intrathalamic contrast but are not part of clinical protocols or extant databases. Here, we introduce Histogram-based polynomial synthesis (HIPS), a fast preprocessing step that synthesizes WMn-like image contrast from standard T1w MRI using a polynomial approximation. HIPS was incorporated into our Thalamus Optimized Multi-Atlas Segmentation (THOMAS) pipeline, developed and optimized for WMn MRI. HIPS-THOMAS was compared to a convolutional neural network (CNN)-based segmentation method and THOMAS modified for T1w images (T1w-THOMAS). The robustness and accuracy of the three methods were tested across different image contrasts, scanner manufacturers, and field strength. HIPS-synthesized images improved intra-thalamic contrast and thalamic boundaries, and their segmentations yielded significantly better mean Dice, lower percentage of volume error, and lower standard deviations compared to both the CNN method and T1w-THOMAS. Finally, using THOMAS, HIPS-synthesized images were as effective as WMn images for identifying thalamic nuclei atrophy in alcohol use disorders subjects relative to healthy controls, with a higher area under the ROC curve compared to T1w-THOMAS (0.79 vs 0.73).
A Contrast Synthesized Thalamic Nuclei Segmentation Scheme using Convolutional Neural Networks
Dec 17, 2020


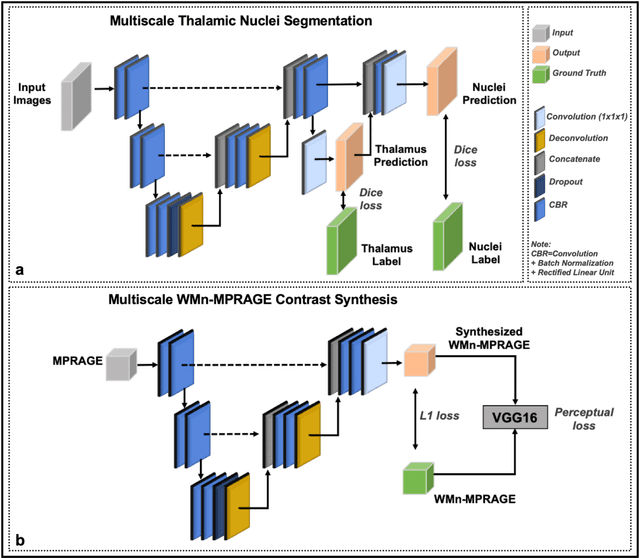
Abstract:Thalamic nuclei have been implicated in several neurological diseases. WMn-MPRAGE images have been shown to provide better intra-thalamic nuclear contrast compared to conventional MPRAGE images but the additional acquisition results in increased examination times. In this work, we investigated 3D Convolutional Neural Network (CNN) based techniques for thalamic nuclei parcellation from conventional MPRAGE images. Two 3D CNNs were developed and compared for thalamic nuclei parcellation using MPRAGE images: a) a native contrast segmentation (NCS) and b) a synthesized contrast segmentation (SCS) using WMn-MPRAGE images synthesized from MPRAGE images. We trained the two segmentation frameworks using MPRAGE images (n=35) and thalamic nuclei labels generated on WMn-MPRAGE images using a multi-atlas based parcellation technique. The segmentation accuracy and clinical utility were evaluated on a cohort comprising of healthy subjects and patients with alcohol use disorder (AUD) (n=45). The SCS network yielded higher Dice scores in the Medial geniculate nucleus (P=.003) and Centromedian nucleus (P=.01) with lower volume differences for Ventral anterior (P=.001) and Ventral posterior lateral (P=.01) nuclei when compared to the NCS network. A Bland-Altman analysis revealed tighter limits of agreement with lower coefficient of variation between true volumes and those predicted by the SCS network. The SCS network demonstrated a significant atrophy in Ventral lateral posterior nucleus in AUD patients compared to healthy age-matched controls (P=0.01), agreeing with previous studies on thalamic atrophy in alcoholism, whereas the NCS network showed spurious atrophy of the Ventral posterior lateral nucleus. CNN-based contrast synthesis prior to segmentation can provide fast and accurate thalamic nuclei segmentation from conventional MPRAGE images.
Robust Automated Thalamic Nuclei Segmentation using a Multi-planar Cascaded Convolutional Neural Network
Dec 16, 2019



Abstract:Purpose: To develop a fast, accurate, and robust convolutional neural network (CNN) based method for segmentation of thalamic nuclei. Methods: A cascaded multi-planar scheme with a modified residual U-Net architecture was used to segment thalamic nuclei on clinical datasets acquired using the white-matter-nulled Magnetization Prepared Rapid Gradient Echo (MPRAGE) sequence. A single network was optimized for healthy controls and disease types (multiple sclerosis, essential tremor) and magnetic field strengths (3T and 7T). Another network was developed to use conventional MPRAGE data. Clinical utility was assessed by comparing a cohort of MS patients to healthy subjects. Results: Segmentation of each thalamus into 12 nuclei was achieved in under 4 minutes. For 7T WMn-MPRAGE, the proposed method outperformed current state-of-the-art with statistically significant improvements in Dice ranging from 1.2% to 5.3% for MS and from 2.6% to 38.8% for ET patients. Comparable accuracy (Dice/VSI) was achieved between 7T and 3T data, attesting to the robustness of the method. For conventional MPRAGE, Dice of > 0.7 was achieved for larger nuclei and > 0.6 for the smaller nuclei. Atrophy of five thalamic nuclei and the whole thalamus was observed for MS patients compared to healthy control subjects, after controlling for intracranial volume and age (p<0.004). Conclusion: The proposed segmentation method is fast, accurate, and generalizes across disease types and field strengths and shows great potential for improving our understanding of thalamic nuclei involvement in neurological diseases and healthy aging. KEYWORDS Deep learning, convolutional neural network, transfer learning, thalamic nuclei segmentation
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge