Mahesh Bharath Keerthivasan
Department of Electrical and Computer Engineering, University of Arizona, Tucson, Arizona, Department of Medical Imaging, University of Arizona, Tucson, Arizona
A Contrast Synthesized Thalamic Nuclei Segmentation Scheme using Convolutional Neural Networks
Dec 17, 2020


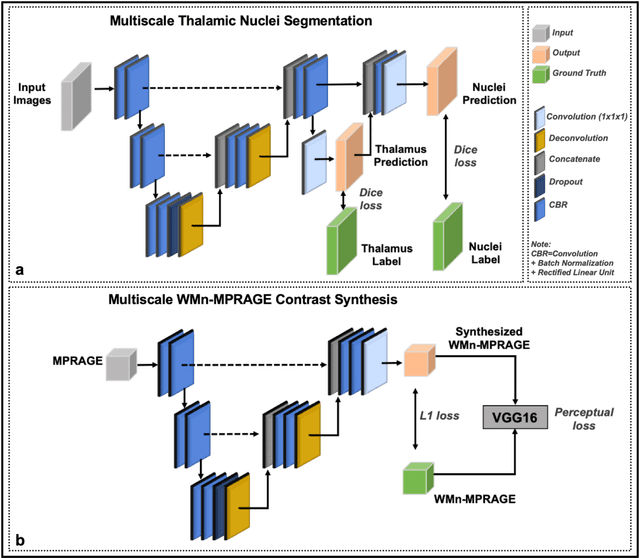
Abstract:Thalamic nuclei have been implicated in several neurological diseases. WMn-MPRAGE images have been shown to provide better intra-thalamic nuclear contrast compared to conventional MPRAGE images but the additional acquisition results in increased examination times. In this work, we investigated 3D Convolutional Neural Network (CNN) based techniques for thalamic nuclei parcellation from conventional MPRAGE images. Two 3D CNNs were developed and compared for thalamic nuclei parcellation using MPRAGE images: a) a native contrast segmentation (NCS) and b) a synthesized contrast segmentation (SCS) using WMn-MPRAGE images synthesized from MPRAGE images. We trained the two segmentation frameworks using MPRAGE images (n=35) and thalamic nuclei labels generated on WMn-MPRAGE images using a multi-atlas based parcellation technique. The segmentation accuracy and clinical utility were evaluated on a cohort comprising of healthy subjects and patients with alcohol use disorder (AUD) (n=45). The SCS network yielded higher Dice scores in the Medial geniculate nucleus (P=.003) and Centromedian nucleus (P=.01) with lower volume differences for Ventral anterior (P=.001) and Ventral posterior lateral (P=.01) nuclei when compared to the NCS network. A Bland-Altman analysis revealed tighter limits of agreement with lower coefficient of variation between true volumes and those predicted by the SCS network. The SCS network demonstrated a significant atrophy in Ventral lateral posterior nucleus in AUD patients compared to healthy age-matched controls (P=0.01), agreeing with previous studies on thalamic atrophy in alcoholism, whereas the NCS network showed spurious atrophy of the Ventral posterior lateral nucleus. CNN-based contrast synthesis prior to segmentation can provide fast and accurate thalamic nuclei segmentation from conventional MPRAGE images.
A Comparison of Deep Learning Convolution Neural Networks for Liver Segmentation in Radial Turbo Spin Echo Images
Apr 13, 2020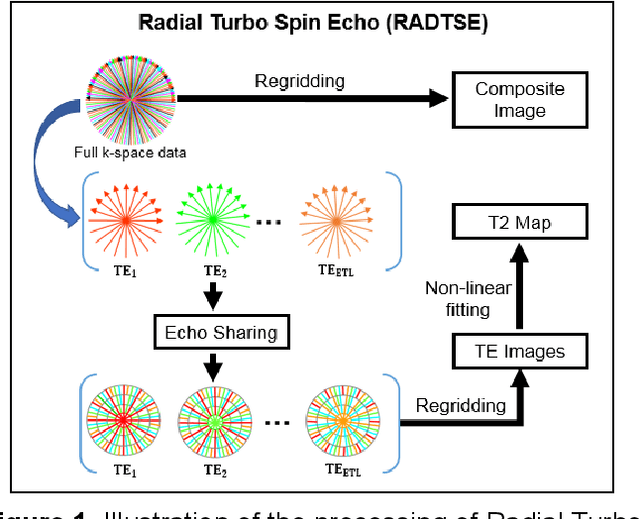
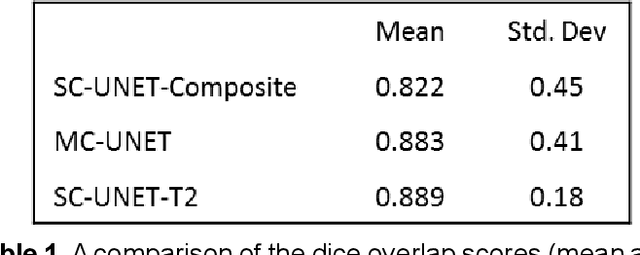
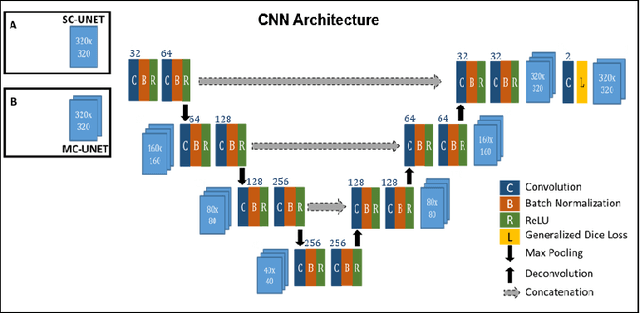
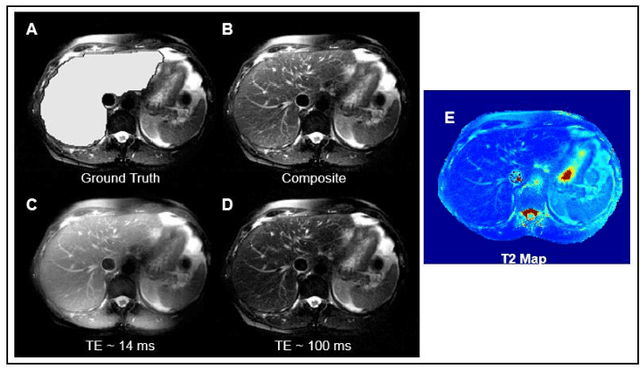
Abstract:Motion-robust 2D Radial Turbo Spin Echo (RADTSE) pulse sequence can provide a high-resolution composite image, T2-weighted images at multiple echo times (TEs), and a quantitative T2 map, all from a single k-space acquisition. In this work, we use a deep-learning convolutional neural network (CNN) for the segmentation of liver in abdominal RADTSE images. A modified UNET architecture with generalized dice loss objective function was implemented. Three 2D CNNs were trained, one for each image type obtained from the RADTSE sequence. On evaluating the performance of the CNNs on the validation set, we found that CNNs trained on TE images or the T2 maps had higher average dice scores than the composite images. This, in turn, implies that the information regarding T2 variation in tissues aids in improving the segmentation performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge