Pierrick Coupé
IM2A
FLAIRBrainSeg: Fine-grained brain segmentation using FLAIR MRI only
Apr 04, 2025Abstract:This paper introduces a novel method for brain segmentation using only FLAIR MRIs, specifically targeting cases where access to other imaging modalities is limited. By leveraging existing automatic segmentation methods, we train a network to approximate segmentations, typically obtained from T1-weighted MRIs. Our method, called FLAIRBrainSeg, produces segmentations of 132 structures and is robust to multiple sclerosis lesions. Experiments on both in-domain and out-of-domain datasets demonstrate that our method outperforms modality-agnostic approaches based on image synthesis, the only currently available alternative for performing brain parcellation using FLAIR MRI alone. This technique holds promise for scenarios where T1-weighted MRIs are unavailable and offers a valuable alternative for clinicians and researchers in need of reliable anatomical segmentation.
Ultra-high resolution multimodal MRI dense labelled holistic brain atlas
Jan 28, 2025



Abstract:In this paper, we introduce holiAtlas, a holistic, multimodal and high-resolution human brain atlas. This atlas covers different levels of details of the human brain anatomy, from the organ to the substructure level, using a new dense labelled protocol generated from the fusion of multiple local protocols at different scales. This atlas has been constructed averaging images and segmentations of 75 healthy subjects from the Human Connectome Project database. Specifically, MR images of T1, T2 and WMn (White Matter nulled) contrasts at 0.125 $mm^{3}$ resolution that were nonlinearly registered and averaged using symmetric group-wise normalisation to construct the atlas. At the finest level, the holiAtlas protocol has 350 different labels derived from 10 different delineation protocols. These labels were grouped at different scales to provide a holistic view of the brain at different levels in a coherent and consistent manner. This multiscale and multimodal atlas can be used for the development of new ultra-high resolution segmentation methods that can potentially leverage the early detection of neurological disorders.
DeepCERES: A Deep learning method for cerebellar lobule segmentation using ultra-high resolution multimodal MRI
Jan 23, 2024Abstract:This paper introduces a novel multimodal and high-resolution human brain cerebellum lobule segmentation method. Unlike current tools that operate at standard resolution ($1 \text{ mm}^{3}$) or using mono-modal data, the proposed method improves cerebellum lobule segmentation through the use of a multimodal and ultra-high resolution ($0.125 \text{ mm}^{3}$) training dataset. To develop the method, first, a database of semi-automatically labelled cerebellum lobules was created to train the proposed method with ultra-high resolution T1 and T2 MR images. Then, an ensemble of deep networks has been designed and developed, allowing the proposed method to excel in the complex cerebellum lobule segmentation task, improving precision while being memory efficient. Notably, our approach deviates from the traditional U-Net model by exploring alternative architectures. We have also integrated deep learning with classical machine learning methods incorporating a priori knowledge from multi-atlas segmentation, which improved precision and robustness. Finally, a new online pipeline, named DeepCERES, has been developed to make available the proposed method to the scientific community requiring as input only a single T1 MR image at standard resolution.
DeepThalamus: A novel deep learning method for automatic segmentation of brain thalamic nuclei from multimodal ultra-high resolution MRI
Jan 15, 2024Abstract:The implication of the thalamus in multiple neurological pathologies makes it a structure of interest for volumetric analysis. In the present work, we have designed and implemented a multimodal volumetric deep neural network for the segmentation of thalamic nuclei at ultra-high resolution (0.125 mm3). Current tools either operate at standard resolution (1 mm3) or use monomodal data. To achieve the proposed objective, first, a database of semiautomatically segmented thalamic nuclei was created using ultra-high resolution T1, T2 and White Matter nulled (WMn) images. Then, a novel Deep learning based strategy was designed to obtain the automatic segmentations and trained to improve its robustness and accuaracy using a semisupervised approach. The proposed method was compared with a related state-of-the-art method showing competitive results both in terms of segmentation quality and efficiency. To make the proposed method fully available to the scientific community, a full pipeline able to work with monomodal standard resolution T1 images is also proposed.
3D Transformer based on deformable patch location for differential diagnosis between Alzheimer's disease and Frontotemporal dementia
Sep 06, 2023



Abstract:Alzheimer's disease and Frontotemporal dementia are common types of neurodegenerative disorders that present overlapping clinical symptoms, making their differential diagnosis very challenging. Numerous efforts have been done for the diagnosis of each disease but the problem of multi-class differential diagnosis has not been actively explored. In recent years, transformer-based models have demonstrated remarkable success in various computer vision tasks. However, their use in disease diagnostic is uncommon due to the limited amount of 3D medical data given the large size of such models. In this paper, we present a novel 3D transformer-based architecture using a deformable patch location module to improve the differential diagnosis of Alzheimer's disease and Frontotemporal dementia. Moreover, to overcome the problem of data scarcity, we propose an efficient combination of various data augmentation techniques, adapted for training transformer-based models on 3D structural magnetic resonance imaging data. Finally, we propose to combine our transformer-based model with a traditional machine learning model using brain structure volumes to better exploit the available data. Our experiments demonstrate the effectiveness of the proposed approach, showing competitive results compared to state-of-the-art methods. Moreover, the deformable patch locations can be visualized, revealing the most relevant brain regions used to establish the diagnosis of each disease.
Brain Structure Ages -- A new biomarker for multi-disease classification
Apr 13, 2023Abstract:Age is an important variable to describe the expected brain's anatomy status across the normal aging trajectory. The deviation from that normative aging trajectory may provide some insights into neurological diseases. In neuroimaging, predicted brain age is widely used to analyze different diseases. However, using only the brain age gap information (\ie the difference between the chronological age and the estimated age) can be not enough informative for disease classification problems. In this paper, we propose to extend the notion of global brain age by estimating brain structure ages using structural magnetic resonance imaging. To this end, an ensemble of deep learning models is first used to estimate a 3D aging map (\ie voxel-wise age estimation). Then, a 3D segmentation mask is used to obtain the final brain structure ages. This biomarker can be used in several situations. First, it enables to accurately estimate the brain age for the purpose of anomaly detection at the population level. In this situation, our approach outperforms several state-of-the-art methods. Second, brain structure ages can be used to compute the deviation from the normal aging process of each brain structure. This feature can be used in a multi-disease classification task for an accurate differential diagnosis at the subject level. Finally, the brain structure age deviations of individuals can be visualized, providing some insights about brain abnormality and helping clinicians in real medical contexts.
Deep Grading based on Collective Artificial Intelligence for AD Diagnosis and Prognosis
Nov 28, 2022Abstract:Accurate diagnosis and prognosis of Alzheimer's disease are crucial to develop new therapies and reduce the associated costs. Recently, with the advances of convolutional neural networks, methods have been proposed to automate these two tasks using structural MRI. However, these methods often suffer from lack of interpretability, generalization, and can be limited in terms of performance. In this paper, we propose a novel deep framework designed to overcome these limitations. Our framework consists of two stages. In the first stage, we propose a deep grading model to extract meaningful features. To enhance the robustness of these features against domain shift, we introduce an innovative collective artificial intelligence strategy for training and evaluating steps. In the second stage, we use a graph convolutional neural network to better capture AD signatures. Our experiments based on 2074 subjects show the competitive performance of our deep framework compared to state-of-the-art methods on different datasets for both AD diagnosis and prognosis.
Deep grading for MRI-based differential diagnosis of Alzheimer's disease and Frontotemporal dementia
Nov 25, 2022Abstract:Alzheimer's disease and Frontotemporal dementia are common forms of neurodegenerative dementia. Behavioral alterations and cognitive impairments are found in the clinical courses of both diseases and their differential diagnosis is sometimes difficult for physicians. Therefore, an accurate tool dedicated to this diagnostic challenge can be valuable in clinical practice. However, current structural imaging methods mainly focus on the detection of each disease but rarely on their differential diagnosis. In this paper, we propose a deep learning based approach for both problems of disease detection and differential diagnosis. We suggest utilizing two types of biomarkers for this application: structure grading and structure atrophy. First, we propose to train a large ensemble of 3D U-Nets to locally determine the anatomical patterns of healthy people, patients with Alzheimer's disease and patients with Frontotemporal dementia using structural MRI as input. The output of the ensemble is a 2-channel disease's coordinate map able to be transformed into a 3D grading map which is easy to interpret for clinicians. This 2-channel map is coupled with a multi-layer perceptron classifier for different classification tasks. Second, we propose to combine our deep learning framework with a traditional machine learning strategy based on volume to improve the model discriminative capacity and robustness. After both cross-validation and external validation, our experiments based on 3319 MRI demonstrated competitive results of our method compared to the state-of-the-art methods for both disease detection and differential diagnosis.
Longitudinal detection of new MS lesions using Deep Learning
Jun 16, 2022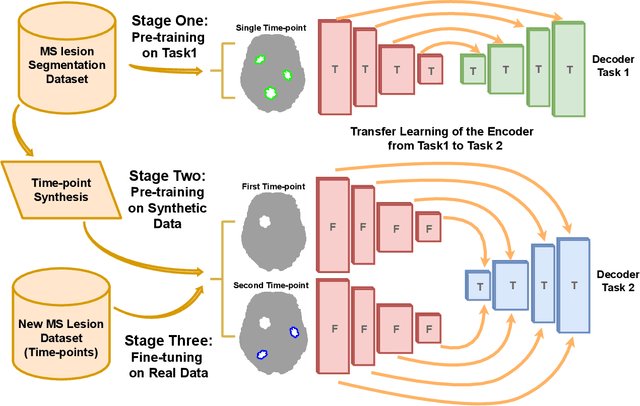
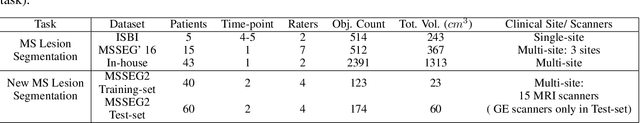
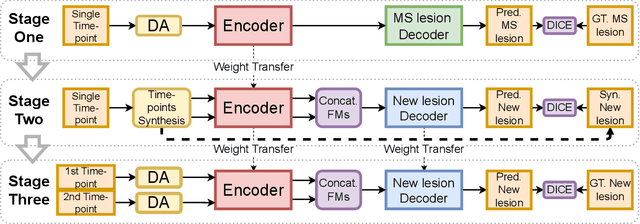
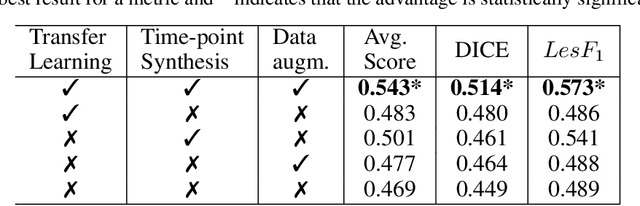
Abstract:The detection of new multiple sclerosis (MS) lesions is an important marker of the evolution of the disease. The applicability of learning-based methods could automate this task efficiently. However, the lack of annotated longitudinal data with new-appearing lesions is a limiting factor for the training of robust and generalizing models. In this work, we describe a deep-learning-based pipeline addressing the challenging task of detecting and segmenting new MS lesions. First, we propose to use transfer-learning from a model trained on a segmentation task using single time-points. Therefore, we exploit knowledge from an easier task and for which more annotated datasets are available. Second, we propose a data synthesis strategy to generate realistic longitudinal time-points with new lesions using single time-point scans. In this way, we pretrain our detection model on large synthetic annotated datasets. Finally, we use a data-augmentation technique designed to simulate data diversity in MRI. By doing that, we increase the size of the available small annotated longitudinal datasets. Our ablation study showed that each contribution lead to an enhancement of the segmentation accuracy. Using the proposed pipeline, we obtained the best score for the segmentation and the detection of new MS lesions in the MSSEG2 MICCAI challenge.
Interpretable differential diagnosis for Alzheimer's disease and Frontotemporal dementia
Jun 15, 2022
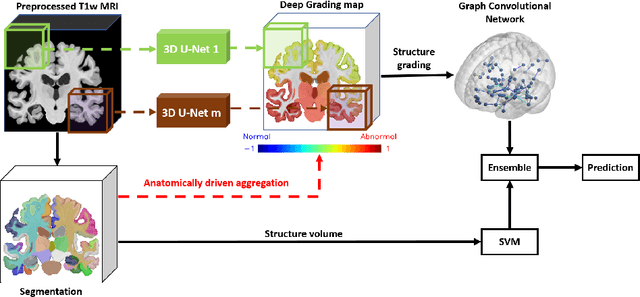
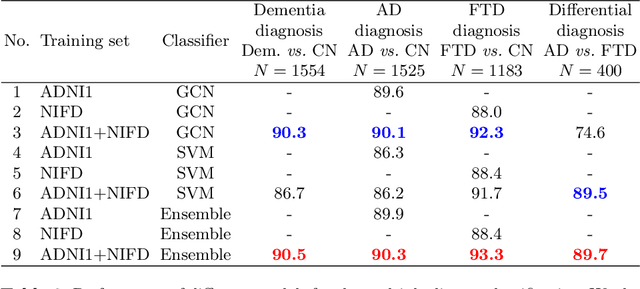
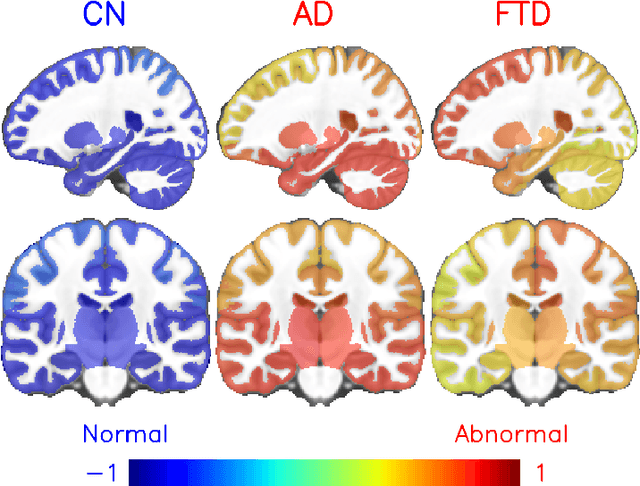
Abstract:Alzheimer's disease and Frontotemporal dementia are two major types of dementia. Their accurate diagnosis and differentiation is crucial for determining specific intervention and treatment. However, differential diagnosis of these two types of dementia remains difficult at the early stage of disease due to similar patterns of clinical symptoms. Therefore, the automatic classification of multiple types of dementia has an important clinical value. So far, this challenge has not been actively explored. Recent development of deep learning in the field of medical image has demonstrated high performance for various classification tasks. In this paper, we propose to take advantage of two types of biomarkers: structure grading and structure atrophy. To this end, we propose first to train a large ensemble of 3D U-Nets to locally discriminate healthy versus dementia anatomical patterns. The result of these models is an interpretable 3D grading map capable of indicating abnormal brain regions. This map can also be exploited in various classification tasks using graph convolutional neural network. Finally, we propose to combine deep grading and atrophy-based classifications to improve dementia type discrimination. The proposed framework showed competitive performance compared to state-of-the-art methods for different tasks of disease detection and differential diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge