Gwenaelle Catheline
Ultra-high resolution multimodal MRI dense labelled holistic brain atlas
Jan 28, 2025



Abstract:In this paper, we introduce holiAtlas, a holistic, multimodal and high-resolution human brain atlas. This atlas covers different levels of details of the human brain anatomy, from the organ to the substructure level, using a new dense labelled protocol generated from the fusion of multiple local protocols at different scales. This atlas has been constructed averaging images and segmentations of 75 healthy subjects from the Human Connectome Project database. Specifically, MR images of T1, T2 and WMn (White Matter nulled) contrasts at 0.125 $mm^{3}$ resolution that were nonlinearly registered and averaged using symmetric group-wise normalisation to construct the atlas. At the finest level, the holiAtlas protocol has 350 different labels derived from 10 different delineation protocols. These labels were grouped at different scales to provide a holistic view of the brain at different levels in a coherent and consistent manner. This multiscale and multimodal atlas can be used for the development of new ultra-high resolution segmentation methods that can potentially leverage the early detection of neurological disorders.
DeepCERES: A Deep learning method for cerebellar lobule segmentation using ultra-high resolution multimodal MRI
Jan 23, 2024Abstract:This paper introduces a novel multimodal and high-resolution human brain cerebellum lobule segmentation method. Unlike current tools that operate at standard resolution ($1 \text{ mm}^{3}$) or using mono-modal data, the proposed method improves cerebellum lobule segmentation through the use of a multimodal and ultra-high resolution ($0.125 \text{ mm}^{3}$) training dataset. To develop the method, first, a database of semi-automatically labelled cerebellum lobules was created to train the proposed method with ultra-high resolution T1 and T2 MR images. Then, an ensemble of deep networks has been designed and developed, allowing the proposed method to excel in the complex cerebellum lobule segmentation task, improving precision while being memory efficient. Notably, our approach deviates from the traditional U-Net model by exploring alternative architectures. We have also integrated deep learning with classical machine learning methods incorporating a priori knowledge from multi-atlas segmentation, which improved precision and robustness. Finally, a new online pipeline, named DeepCERES, has been developed to make available the proposed method to the scientific community requiring as input only a single T1 MR image at standard resolution.
3D Inception-based CNN with sMRI and MD-DTI data fusion for Alzheimer's Disease diagnostics
Jul 17, 2018
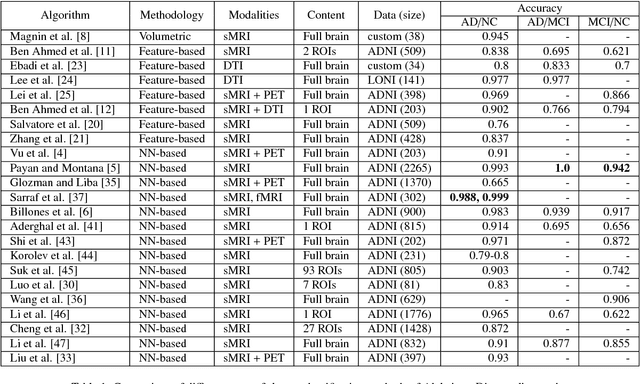
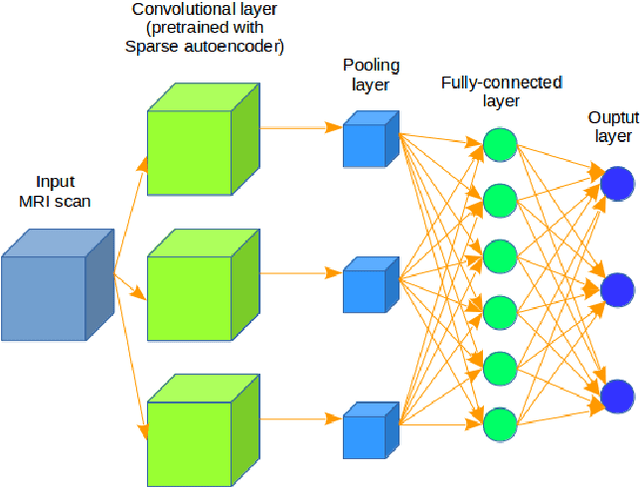
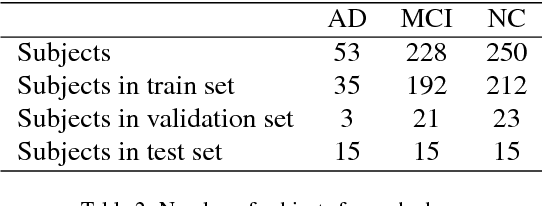
Abstract:In the last decade, computer-aided early diagnostics of Alzheimer's Disease (AD) and its prodromal form, Mild Cognitive Impairment (MCI), has been the subject of extensive research. Some recent studies have shown promising results in the AD and MCI determination using structural and functional Magnetic Resonance Imaging (sMRI, fMRI), Positron Emission Tomography (PET) and Diffusion Tensor Imaging (DTI) modalities. Furthermore, fusion of imaging modalities in a supervised machine learning framework has shown promising direction of research. In this paper we first review major trends in automatic classification methods such as feature extraction based methods as well as deep learning approaches in medical image analysis applied to the field of Alzheimer's Disease diagnostics. Then we propose our own design of a 3D Inception-based Convolutional Neural Network (CNN) for Alzheimer's Disease diagnostics. The network is designed with an emphasis on the interior resource utilization and uses sMRI and DTI modalities fusion on hippocampal ROI. The comparison with the conventional AlexNet-based network using data from the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset (http://adni.loni.usc.edu) demonstrates significantly better performance of the proposed 3D Inception-based CNN.
3D CNN-based classification using sMRI and MD-DTI images for Alzheimer disease studies
Jan 18, 2018
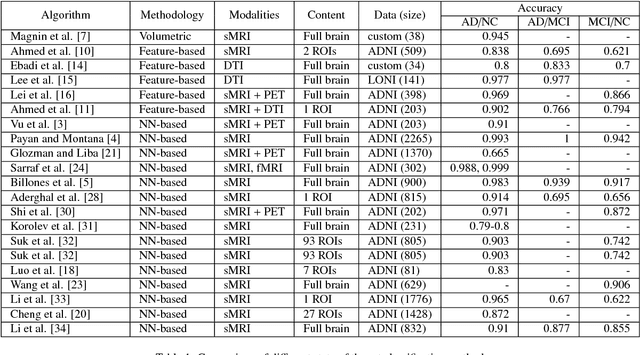
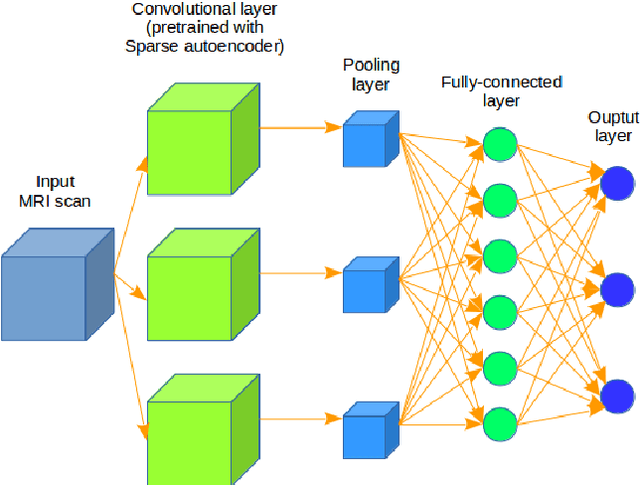
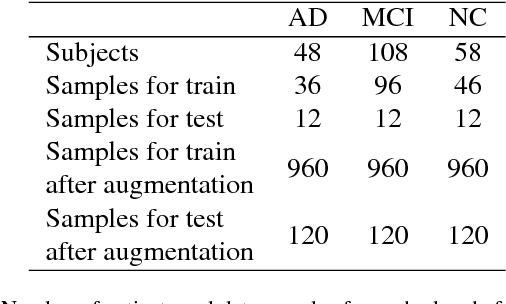
Abstract:Computer-aided early diagnosis of Alzheimers Disease (AD) and its prodromal form, Mild Cognitive Impairment (MCI), has been the subject of extensive research in recent years. Some recent studies have shown promising results in the AD and MCI determination using structural and functional Magnetic Resonance Imaging (sMRI, fMRI), Positron Emission Tomography (PET) and Diffusion Tensor Imaging (DTI) modalities. Furthermore, fusion of imaging modalities in a supervised machine learning framework has shown promising direction of research. In this paper we first review major trends in automatic classification methods such as feature extraction based methods as well as deep learning approaches in medical image analysis applied to the field of Alzheimer's Disease diagnostics. Then we propose our own algorithm for Alzheimer's Disease diagnostics based on a convolutional neural network and sMRI and DTI modalities fusion on hippocampal ROI using data from the Alzheimers Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). Comparison with a single modality approach shows promising results. We also propose our own method of data augmentation for balancing classes of different size and analyze the impact of the ROI size on the classification results as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge