Thomas Tourdias
FLAIRBrainSeg: Fine-grained brain segmentation using FLAIR MRI only
Apr 04, 2025Abstract:This paper introduces a novel method for brain segmentation using only FLAIR MRIs, specifically targeting cases where access to other imaging modalities is limited. By leveraging existing automatic segmentation methods, we train a network to approximate segmentations, typically obtained from T1-weighted MRIs. Our method, called FLAIRBrainSeg, produces segmentations of 132 structures and is robust to multiple sclerosis lesions. Experiments on both in-domain and out-of-domain datasets demonstrate that our method outperforms modality-agnostic approaches based on image synthesis, the only currently available alternative for performing brain parcellation using FLAIR MRI alone. This technique holds promise for scenarios where T1-weighted MRIs are unavailable and offers a valuable alternative for clinicians and researchers in need of reliable anatomical segmentation.
Ultra-high resolution multimodal MRI dense labelled holistic brain atlas
Jan 28, 2025



Abstract:In this paper, we introduce holiAtlas, a holistic, multimodal and high-resolution human brain atlas. This atlas covers different levels of details of the human brain anatomy, from the organ to the substructure level, using a new dense labelled protocol generated from the fusion of multiple local protocols at different scales. This atlas has been constructed averaging images and segmentations of 75 healthy subjects from the Human Connectome Project database. Specifically, MR images of T1, T2 and WMn (White Matter nulled) contrasts at 0.125 $mm^{3}$ resolution that were nonlinearly registered and averaged using symmetric group-wise normalisation to construct the atlas. At the finest level, the holiAtlas protocol has 350 different labels derived from 10 different delineation protocols. These labels were grouped at different scales to provide a holistic view of the brain at different levels in a coherent and consistent manner. This multiscale and multimodal atlas can be used for the development of new ultra-high resolution segmentation methods that can potentially leverage the early detection of neurological disorders.
DeepThalamus: A novel deep learning method for automatic segmentation of brain thalamic nuclei from multimodal ultra-high resolution MRI
Jan 15, 2024Abstract:The implication of the thalamus in multiple neurological pathologies makes it a structure of interest for volumetric analysis. In the present work, we have designed and implemented a multimodal volumetric deep neural network for the segmentation of thalamic nuclei at ultra-high resolution (0.125 mm3). Current tools either operate at standard resolution (1 mm3) or use monomodal data. To achieve the proposed objective, first, a database of semiautomatically segmented thalamic nuclei was created using ultra-high resolution T1, T2 and White Matter nulled (WMn) images. Then, a novel Deep learning based strategy was designed to obtain the automatic segmentations and trained to improve its robustness and accuaracy using a semisupervised approach. The proposed method was compared with a related state-of-the-art method showing competitive results both in terms of segmentation quality and efficiency. To make the proposed method fully available to the scientific community, a full pipeline able to work with monomodal standard resolution T1 images is also proposed.
Towards broader generalization of deep learning methods for multiple sclerosis lesion segmentation
Dec 14, 2020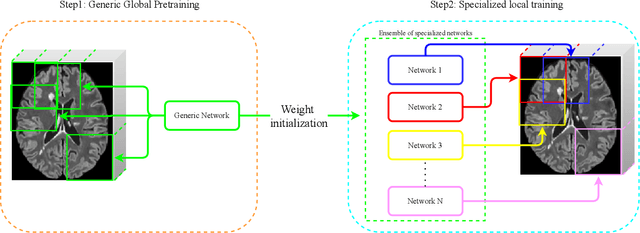
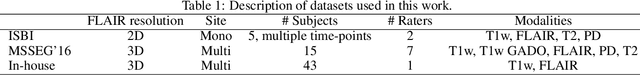
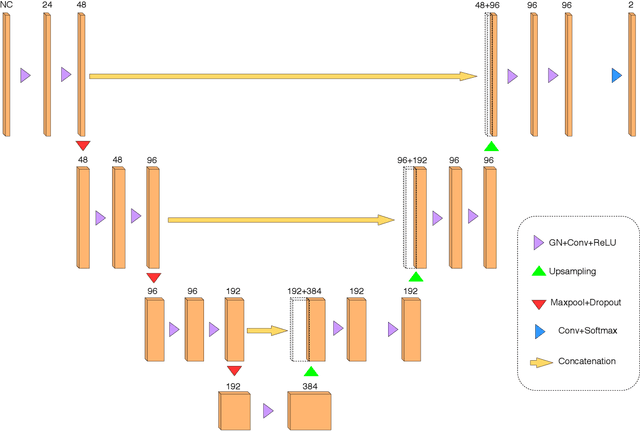
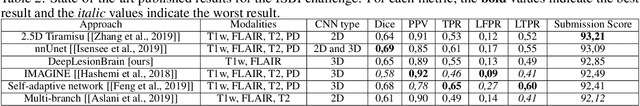
Abstract:Recently, segmentation methods based on Convolutional Neural Networks (CNNs) showed promising performance in automatic Multiple Sclerosis (MS) lesions segmentation. These techniques have even outperformed human experts in controlled evaluation condition. However state-of-the-art approaches trained to perform well on highly-controlled datasets fail to generalize on clinical data from unseen datasets. Instead of proposing another improvement of the segmentation accuracy, we propose a novel method robust to domain shift and performing well on unseen datasets, called DeepLesionBrain (DLB). This generalization property results from three main contributions. First, DLB is based on a large ensemble of compact 3D CNNs. This ensemble strategy ensures a robust prediction despite the risk of generalization failure of some individual networks. Second, DLB includes a new image quality data augmentation to reduce dependency to training data specificity (e.g., acquisition protocol). Finally, to learn a more generalizable representation of MS lesions, we propose a hierarchical specialization learning (HSL). HSL is performed by pre-training a generic network over the whole brain, before using its weights as initialization to locally specialized networks. By this end, DLB learns both generic features extracted at global image level and specific features extracted at local image level. At the time of publishing this paper, DLB is among the Top 3 performing published methods on ISBI Challenge while using only half of the available modalities. DLB generalization has also been compared to other state-of-the-art approaches, during cross-dataset experiments on MSSEG'16, ISBI challenge, and in-house datasets. DLB improves the segmentation performance and generalization over classical techniques, and thus proposes a robust approach better suited for clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge