Fernando Aparici
Ultra-high resolution multimodal MRI dense labelled holistic brain atlas
Jan 28, 2025



Abstract:In this paper, we introduce holiAtlas, a holistic, multimodal and high-resolution human brain atlas. This atlas covers different levels of details of the human brain anatomy, from the organ to the substructure level, using a new dense labelled protocol generated from the fusion of multiple local protocols at different scales. This atlas has been constructed averaging images and segmentations of 75 healthy subjects from the Human Connectome Project database. Specifically, MR images of T1, T2 and WMn (White Matter nulled) contrasts at 0.125 $mm^{3}$ resolution that were nonlinearly registered and averaged using symmetric group-wise normalisation to construct the atlas. At the finest level, the holiAtlas protocol has 350 different labels derived from 10 different delineation protocols. These labels were grouped at different scales to provide a holistic view of the brain at different levels in a coherent and consistent manner. This multiscale and multimodal atlas can be used for the development of new ultra-high resolution segmentation methods that can potentially leverage the early detection of neurological disorders.
DeepCERES: A Deep learning method for cerebellar lobule segmentation using ultra-high resolution multimodal MRI
Jan 23, 2024Abstract:This paper introduces a novel multimodal and high-resolution human brain cerebellum lobule segmentation method. Unlike current tools that operate at standard resolution ($1 \text{ mm}^{3}$) or using mono-modal data, the proposed method improves cerebellum lobule segmentation through the use of a multimodal and ultra-high resolution ($0.125 \text{ mm}^{3}$) training dataset. To develop the method, first, a database of semi-automatically labelled cerebellum lobules was created to train the proposed method with ultra-high resolution T1 and T2 MR images. Then, an ensemble of deep networks has been designed and developed, allowing the proposed method to excel in the complex cerebellum lobule segmentation task, improving precision while being memory efficient. Notably, our approach deviates from the traditional U-Net model by exploring alternative architectures. We have also integrated deep learning with classical machine learning methods incorporating a priori knowledge from multi-atlas segmentation, which improved precision and robustness. Finally, a new online pipeline, named DeepCERES, has been developed to make available the proposed method to the scientific community requiring as input only a single T1 MR image at standard resolution.
DeepThalamus: A novel deep learning method for automatic segmentation of brain thalamic nuclei from multimodal ultra-high resolution MRI
Jan 15, 2024Abstract:The implication of the thalamus in multiple neurological pathologies makes it a structure of interest for volumetric analysis. In the present work, we have designed and implemented a multimodal volumetric deep neural network for the segmentation of thalamic nuclei at ultra-high resolution (0.125 mm3). Current tools either operate at standard resolution (1 mm3) or use monomodal data. To achieve the proposed objective, first, a database of semiautomatically segmented thalamic nuclei was created using ultra-high resolution T1, T2 and White Matter nulled (WMn) images. Then, a novel Deep learning based strategy was designed to obtain the automatic segmentations and trained to improve its robustness and accuaracy using a semisupervised approach. The proposed method was compared with a related state-of-the-art method showing competitive results both in terms of segmentation quality and efficiency. To make the proposed method fully available to the scientific community, a full pipeline able to work with monomodal standard resolution T1 images is also proposed.
vol2Brain: A new online Pipeline for whole Brain MRI analysis
Feb 08, 2022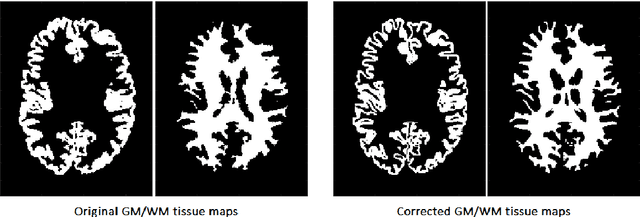
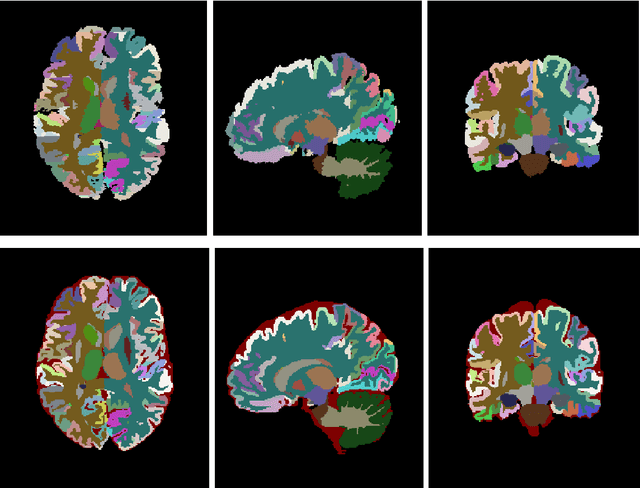

Abstract:Automatic and reliable quantitative tools for MR brain image analysis are a very valuable resources for both clinical and research environments. In the last years, this field has experienced many advances with successful techniques based on label fusion and more recently deep learning. However, few of them have been specifically designed to provide a dense anatomical labelling at multiscale level and to deal with brain anatomical alterations such as white matter lesions. In this work, we present a fully automatic pipeline (vol2Brain) for whole brain segmentation and analysis which densely labels (N>100) the brain while being robust to the presence of white matter lesions. This new pipeline is an evolution of our previous volBrain pipeline that extends significantly the number of regions that can be analyzed. Our proposed method is based on a fast multiscale multi-atlas label fusion technology with systematic error correction able to provide accurate volumetric information in few minutes. We have deployed our new pipeline within our platform volBrain (www.volbrain.upv.es) which has been already demonstrated to be an efficient and effective manner to share our technology with users worldwide
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge