José V Manjon
Longitudinal detection of new MS lesions using Deep Learning
Jun 16, 2022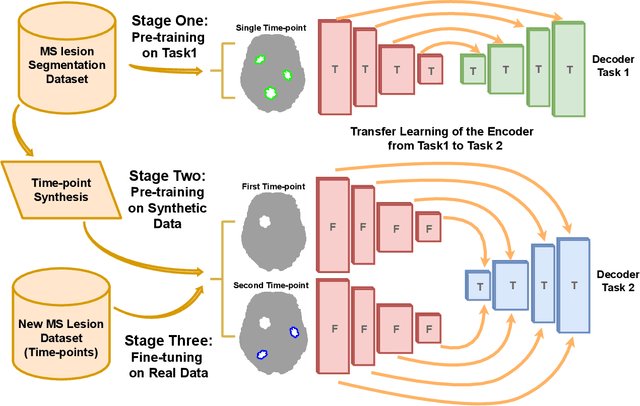
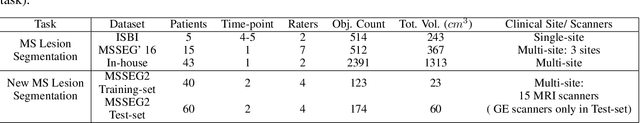
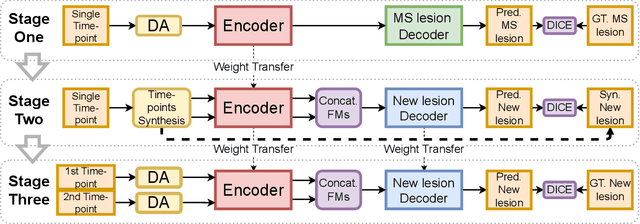
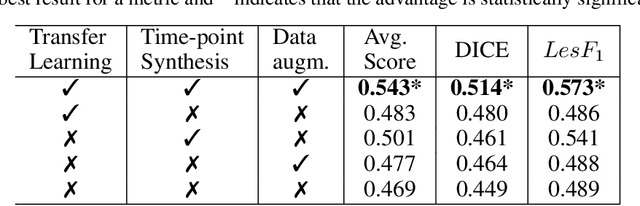
Abstract:The detection of new multiple sclerosis (MS) lesions is an important marker of the evolution of the disease. The applicability of learning-based methods could automate this task efficiently. However, the lack of annotated longitudinal data with new-appearing lesions is a limiting factor for the training of robust and generalizing models. In this work, we describe a deep-learning-based pipeline addressing the challenging task of detecting and segmenting new MS lesions. First, we propose to use transfer-learning from a model trained on a segmentation task using single time-points. Therefore, we exploit knowledge from an easier task and for which more annotated datasets are available. Second, we propose a data synthesis strategy to generate realistic longitudinal time-points with new lesions using single time-point scans. In this way, we pretrain our detection model on large synthetic annotated datasets. Finally, we use a data-augmentation technique designed to simulate data diversity in MRI. By doing that, we increase the size of the available small annotated longitudinal datasets. Our ablation study showed that each contribution lead to an enhancement of the segmentation accuracy. Using the proposed pipeline, we obtained the best score for the segmentation and the detection of new MS lesions in the MSSEG2 MICCAI challenge.
POPCORN: Progressive Pseudo-labeling with Consistency Regularization and Neighboring
Sep 13, 2021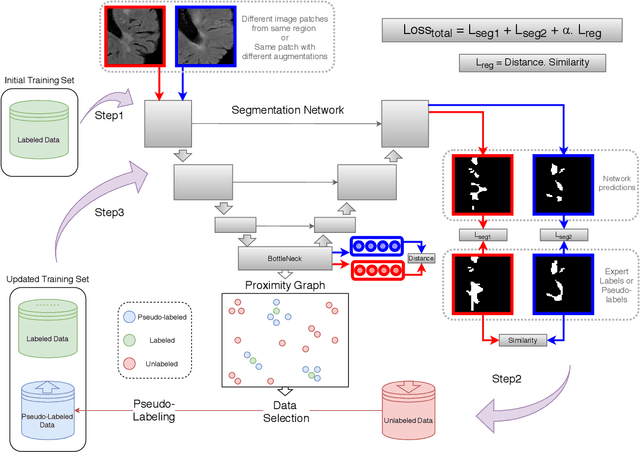
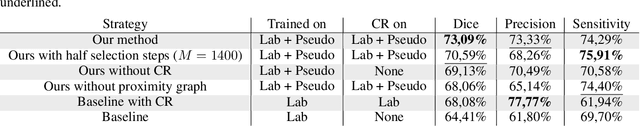
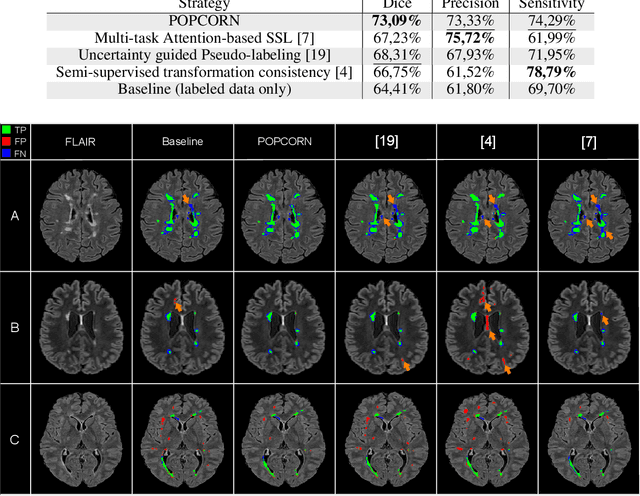
Abstract:Semi-supervised learning (SSL) uses unlabeled data to compensate for the scarcity of annotated images and the lack of method generalization to unseen domains, two usual problems in medical segmentation tasks. In this work, we propose POPCORN, a novel method combining consistency regularization and pseudo-labeling designed for image segmentation. The proposed framework uses high-level regularization to constrain our segmentation model to use similar latent features for images with similar segmentations. POPCORN estimates a proximity graph to select data from easiest ones to more difficult ones, in order to ensure accurate pseudo-labeling and to limit confirmation bias. Applied to multiple sclerosis lesion segmentation, our method demonstrates competitive results compared to other state-of-the-art SSL strategies.
Towards broader generalization of deep learning methods for multiple sclerosis lesion segmentation
Dec 14, 2020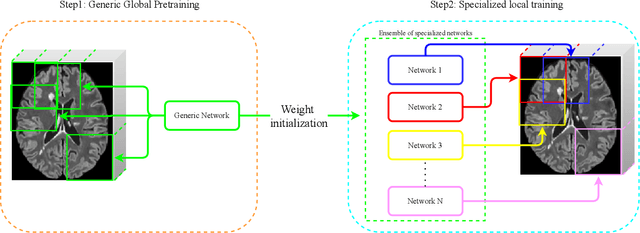
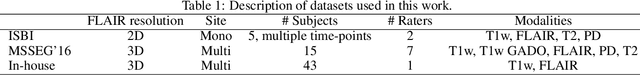
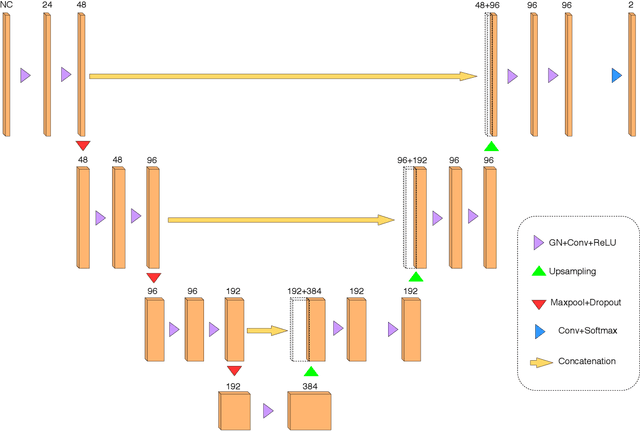
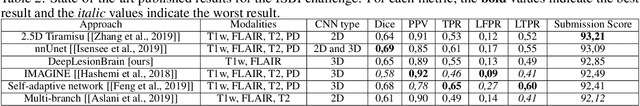
Abstract:Recently, segmentation methods based on Convolutional Neural Networks (CNNs) showed promising performance in automatic Multiple Sclerosis (MS) lesions segmentation. These techniques have even outperformed human experts in controlled evaluation condition. However state-of-the-art approaches trained to perform well on highly-controlled datasets fail to generalize on clinical data from unseen datasets. Instead of proposing another improvement of the segmentation accuracy, we propose a novel method robust to domain shift and performing well on unseen datasets, called DeepLesionBrain (DLB). This generalization property results from three main contributions. First, DLB is based on a large ensemble of compact 3D CNNs. This ensemble strategy ensures a robust prediction despite the risk of generalization failure of some individual networks. Second, DLB includes a new image quality data augmentation to reduce dependency to training data specificity (e.g., acquisition protocol). Finally, to learn a more generalizable representation of MS lesions, we propose a hierarchical specialization learning (HSL). HSL is performed by pre-training a generic network over the whole brain, before using its weights as initialization to locally specialized networks. By this end, DLB learns both generic features extracted at global image level and specific features extracted at local image level. At the time of publishing this paper, DLB is among the Top 3 performing published methods on ISBI Challenge while using only half of the available modalities. DLB generalization has also been compared to other state-of-the-art approaches, during cross-dataset experiments on MSSEG'16, ISBI challenge, and in-house datasets. DLB improves the segmentation performance and generalization over classical techniques, and thus proposes a robust approach better suited for clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge