Michaël Clément
Superpixel Anything: A general object-based framework for accurate yet regular superpixel segmentation
Sep 16, 2025Abstract:Superpixels are widely used in computer vision to simplify image representation and reduce computational complexity. While traditional methods rely on low-level features, deep learning-based approaches leverage high-level features but also tend to sacrifice regularity of superpixels to capture complex objects, leading to accurate but less interpretable segmentations. In this work, we introduce SPAM (SuperPixel Anything Model), a versatile framework for segmenting images into accurate yet regular superpixels. We train a model to extract image features for superpixel generation, and at inference, we leverage a large-scale pretrained model for semantic-agnostic segmentation to ensure that superpixels align with object masks. SPAM can handle any prior high-level segmentation, resolving uncertainty regions, and is able to interactively focus on specific objects. Comprehensive experiments demonstrate that SPAM qualitatively and quantitatively outperforms state-of-the-art methods on segmentation tasks, making it a valuable and robust tool for various applications. Code and pre-trained models are available here: https://github.com/waldo-j/spam.
Superpixel Segmentation: A Long-Lasting Ill-Posed Problem
Nov 10, 2024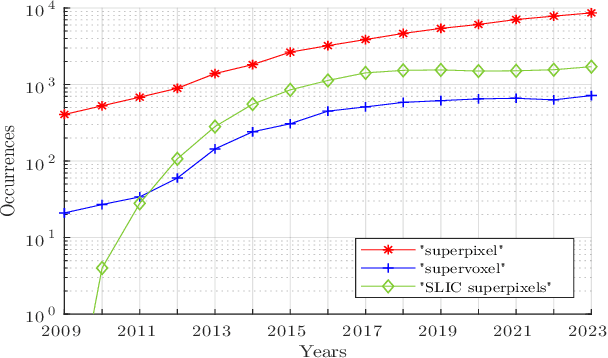
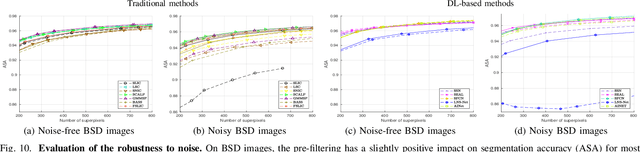
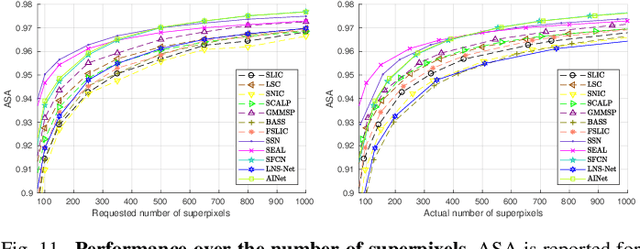

Abstract:For many years, image over-segmentation into superpixels has been essential to computer vision pipelines, by creating homogeneous and identifiable regions of similar sizes. Such constrained segmentation problem would require a clear definition and specific evaluation criteria. However, the validation framework for superpixel methods, typically viewed as standard object segmentation, has rarely been thoroughly studied. In this work, we first take a step back to show that superpixel segmentation is fundamentally an ill-posed problem, due to the implicit regularity constraint on the shape and size of superpixels. We also demonstrate through a novel comprehensive study that the literature suffers from only evaluating certain aspects, sometimes incorrectly and with inappropriate metrics. Concurrently, recent deep learning-based superpixel methods mainly focus on the object segmentation task at the expense of regularity. In this ill-posed context, we show that we can achieve competitive results using a recent architecture like the Segment Anything Model (SAM), without dedicated training for the superpixel segmentation task. This leads to rethinking superpixel segmentation and the necessary properties depending on the targeted downstream task.
Deep Spherical Superpixels
Jul 24, 2024Abstract:Over the years, the use of superpixel segmentation has become very popular in various applications, serving as a preprocessing step to reduce data size by adapting to the content of the image, regardless of its semantic content. While the superpixel segmentation of standard planar images, captured with a 90{\deg} field of view, has been extensively studied, there has been limited focus on dedicated methods to omnidirectional or spherical images, captured with a 360{\deg} field of view. In this study, we introduce the first deep learning-based superpixel segmentation approach tailored for omnidirectional images called DSS (for Deep Spherical Superpixels). Our methodology leverages on spherical CNN architectures and the differentiable K-means clustering paradigm for superpixels, to generate superpixels that follow the spherical geometry. Additionally, we propose to use data augmentation techniques specifically designed for 360{\deg} images, enabling our model to efficiently learn from a limited set of annotated omnidirectional data. Our extensive validation across two datasets demonstrates that taking into account the inherent circular geometry of such images into our framework improves the segmentation performance over traditional and deep learning-based superpixel methods. Our code is available online.
3D Transformer based on deformable patch location for differential diagnosis between Alzheimer's disease and Frontotemporal dementia
Sep 06, 2023



Abstract:Alzheimer's disease and Frontotemporal dementia are common types of neurodegenerative disorders that present overlapping clinical symptoms, making their differential diagnosis very challenging. Numerous efforts have been done for the diagnosis of each disease but the problem of multi-class differential diagnosis has not been actively explored. In recent years, transformer-based models have demonstrated remarkable success in various computer vision tasks. However, their use in disease diagnostic is uncommon due to the limited amount of 3D medical data given the large size of such models. In this paper, we present a novel 3D transformer-based architecture using a deformable patch location module to improve the differential diagnosis of Alzheimer's disease and Frontotemporal dementia. Moreover, to overcome the problem of data scarcity, we propose an efficient combination of various data augmentation techniques, adapted for training transformer-based models on 3D structural magnetic resonance imaging data. Finally, we propose to combine our transformer-based model with a traditional machine learning model using brain structure volumes to better exploit the available data. Our experiments demonstrate the effectiveness of the proposed approach, showing competitive results compared to state-of-the-art methods. Moreover, the deformable patch locations can be visualized, revealing the most relevant brain regions used to establish the diagnosis of each disease.
Brain Structure Ages -- A new biomarker for multi-disease classification
Apr 13, 2023Abstract:Age is an important variable to describe the expected brain's anatomy status across the normal aging trajectory. The deviation from that normative aging trajectory may provide some insights into neurological diseases. In neuroimaging, predicted brain age is widely used to analyze different diseases. However, using only the brain age gap information (\ie the difference between the chronological age and the estimated age) can be not enough informative for disease classification problems. In this paper, we propose to extend the notion of global brain age by estimating brain structure ages using structural magnetic resonance imaging. To this end, an ensemble of deep learning models is first used to estimate a 3D aging map (\ie voxel-wise age estimation). Then, a 3D segmentation mask is used to obtain the final brain structure ages. This biomarker can be used in several situations. First, it enables to accurately estimate the brain age for the purpose of anomaly detection at the population level. In this situation, our approach outperforms several state-of-the-art methods. Second, brain structure ages can be used to compute the deviation from the normal aging process of each brain structure. This feature can be used in a multi-disease classification task for an accurate differential diagnosis at the subject level. Finally, the brain structure age deviations of individuals can be visualized, providing some insights about brain abnormality and helping clinicians in real medical contexts.
Deep Grading based on Collective Artificial Intelligence for AD Diagnosis and Prognosis
Nov 28, 2022Abstract:Accurate diagnosis and prognosis of Alzheimer's disease are crucial to develop new therapies and reduce the associated costs. Recently, with the advances of convolutional neural networks, methods have been proposed to automate these two tasks using structural MRI. However, these methods often suffer from lack of interpretability, generalization, and can be limited in terms of performance. In this paper, we propose a novel deep framework designed to overcome these limitations. Our framework consists of two stages. In the first stage, we propose a deep grading model to extract meaningful features. To enhance the robustness of these features against domain shift, we introduce an innovative collective artificial intelligence strategy for training and evaluating steps. In the second stage, we use a graph convolutional neural network to better capture AD signatures. Our experiments based on 2074 subjects show the competitive performance of our deep framework compared to state-of-the-art methods on different datasets for both AD diagnosis and prognosis.
Deep grading for MRI-based differential diagnosis of Alzheimer's disease and Frontotemporal dementia
Nov 25, 2022Abstract:Alzheimer's disease and Frontotemporal dementia are common forms of neurodegenerative dementia. Behavioral alterations and cognitive impairments are found in the clinical courses of both diseases and their differential diagnosis is sometimes difficult for physicians. Therefore, an accurate tool dedicated to this diagnostic challenge can be valuable in clinical practice. However, current structural imaging methods mainly focus on the detection of each disease but rarely on their differential diagnosis. In this paper, we propose a deep learning based approach for both problems of disease detection and differential diagnosis. We suggest utilizing two types of biomarkers for this application: structure grading and structure atrophy. First, we propose to train a large ensemble of 3D U-Nets to locally determine the anatomical patterns of healthy people, patients with Alzheimer's disease and patients with Frontotemporal dementia using structural MRI as input. The output of the ensemble is a 2-channel disease's coordinate map able to be transformed into a 3D grading map which is easy to interpret for clinicians. This 2-channel map is coupled with a multi-layer perceptron classifier for different classification tasks. Second, we propose to combine our deep learning framework with a traditional machine learning strategy based on volume to improve the model discriminative capacity and robustness. After both cross-validation and external validation, our experiments based on 3319 MRI demonstrated competitive results of our method compared to the state-of-the-art methods for both disease detection and differential diagnosis.
Interpretable differential diagnosis for Alzheimer's disease and Frontotemporal dementia
Jun 15, 2022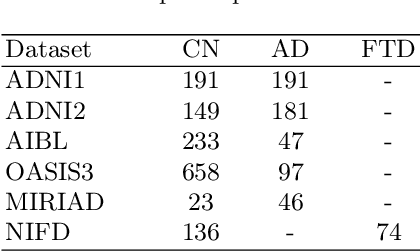
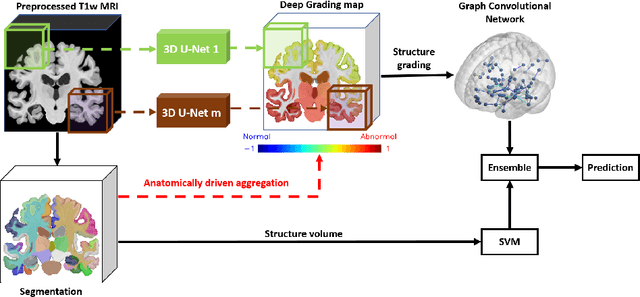
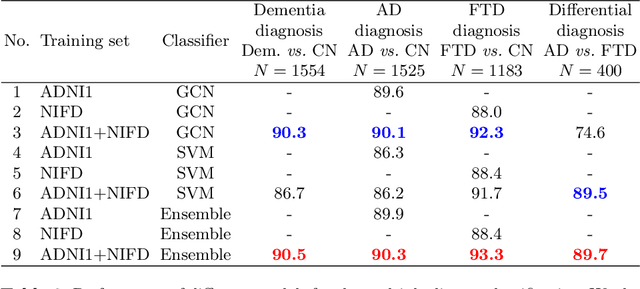
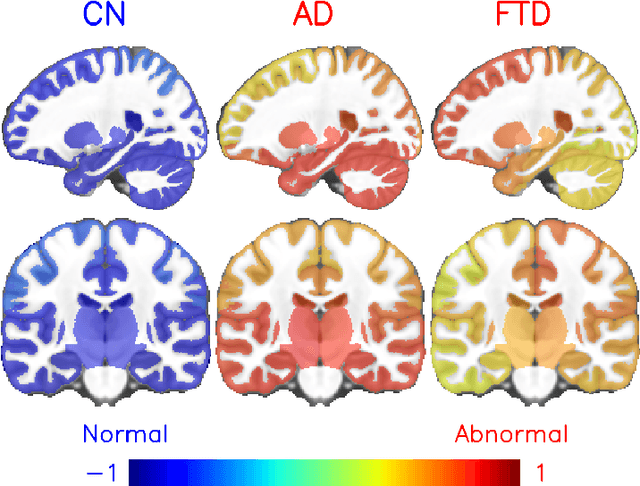
Abstract:Alzheimer's disease and Frontotemporal dementia are two major types of dementia. Their accurate diagnosis and differentiation is crucial for determining specific intervention and treatment. However, differential diagnosis of these two types of dementia remains difficult at the early stage of disease due to similar patterns of clinical symptoms. Therefore, the automatic classification of multiple types of dementia has an important clinical value. So far, this challenge has not been actively explored. Recent development of deep learning in the field of medical image has demonstrated high performance for various classification tasks. In this paper, we propose to take advantage of two types of biomarkers: structure grading and structure atrophy. To this end, we propose first to train a large ensemble of 3D U-Nets to locally discriminate healthy versus dementia anatomical patterns. The result of these models is an interpretable 3D grading map capable of indicating abnormal brain regions. This map can also be exploited in various classification tasks using graph convolutional neural network. Finally, we propose to combine deep grading and atrophy-based classifications to improve dementia type discrimination. The proposed framework showed competitive performance compared to state-of-the-art methods for different tasks of disease detection and differential diagnosis.
Towards better Interpretable and Generalizable AD detection using Collective Artificial Intelligence
Jun 07, 2022

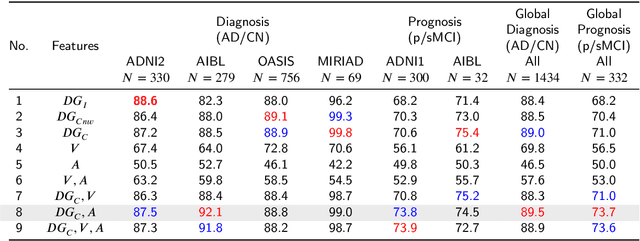

Abstract:Accurate diagnosis and prognosis of Alzheimer's disease are crucial for developing new therapies and reducing the associated costs. Recently, with the advances of convolutional neural networks, deep learning methods have been proposed to automate these two tasks using structural MRI. However, these methods often suffer from a lack of interpretability and generalization and have limited prognosis performance. In this paper, we propose a novel deep framework designed to overcome these limitations. Our pipeline consists of two stages. In the first stage, 125 3D U-Nets are used to estimate voxelwise grade scores over the whole brain. The resulting 3D maps are then fused to construct an interpretable 3D grading map indicating the disease severity at the structure level. As a consequence, clinicians can use this map to detect the brain structures affected by the disease. In the second stage, the grading map and subject's age are used to perform classification with a graph convolutional neural network. Experimental results based on 2106 subjects demonstrated competitive performance of our deep framework compared to state-of-the-art methods on different datasets for both AD diagnosis and prognosis. Moreover, we found that using a large number of U-Nets processing different overlapping brain areas improved the generalization capacity of the proposed methods.
Analysis of Different Losses for Deep Learning Image Colorization
Apr 06, 2022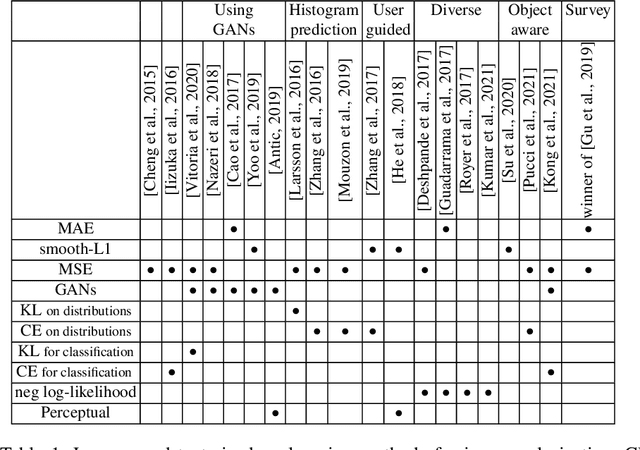
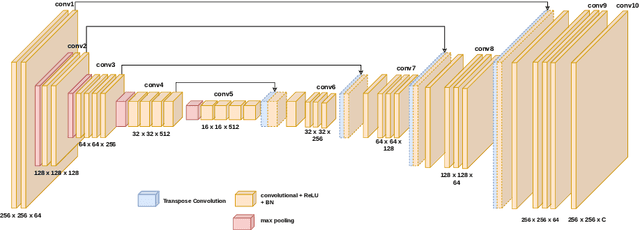
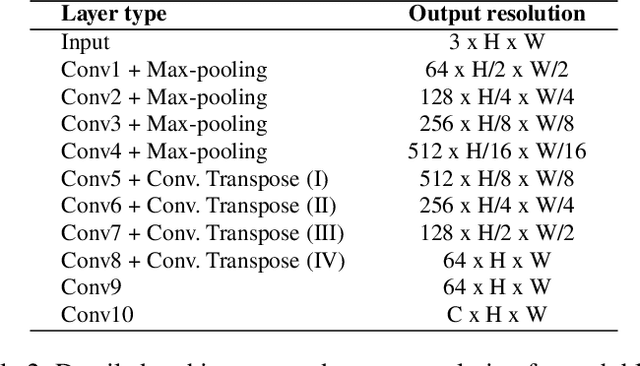

Abstract:Image colorization aims to add color information to a grayscale image in a realistic way. Recent methods mostly rely on deep learning strategies. While learning to automatically colorize an image, one can define well-suited objective functions related to the desired color output. Some of them are based on a specific type of error between the predicted image and ground truth one, while other losses rely on the comparison of perceptual properties. But, is the choice of the objective function that crucial, i.e., does it play an important role in the results? In this chapter, we aim to answer this question by analyzing the impact of the loss function on the estimated colorization results. To that goal, we review the different losses and evaluation metrics that are used in the literature. We then train a baseline network with several of the reviewed objective functions: classic L1 and L2 losses, as well as more complex combinations such as Wasserstein GAN and VGG-based LPIPS loss. Quantitative results show that the models trained with VGG-based LPIPS provide overall slightly better results for most evaluation metrics. Qualitative results exhibit more vivid colors when with Wasserstein GAN plus the L2 loss or again with the VGG-based LPIPS. Finally, the convenience of quantitative user studies is also discussed to overcome the difficulty of properly assessing on colorized images, notably for the case of old archive photographs where no ground truth is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge