Allen Zhang
CG-RAG: Research Question Answering by Citation Graph Retrieval-Augmented LLMs
Jan 25, 2025



Abstract:Research question answering requires accurate retrieval and contextual understanding of scientific literature. However, current Retrieval-Augmented Generation (RAG) methods often struggle to balance complex document relationships with precise information retrieval. In this paper, we introduce Contextualized Graph Retrieval-Augmented Generation (CG-RAG), a novel framework that integrates sparse and dense retrieval signals within graph structures to enhance retrieval efficiency and subsequently improve generation quality for research question answering. First, we propose a contextual graph representation for citation graphs, effectively capturing both explicit and implicit connections within and across documents. Next, we introduce Lexical-Semantic Graph Retrieval (LeSeGR), which seamlessly integrates sparse and dense retrieval signals with graph encoding. It bridges the gap between lexical precision and semantic understanding in citation graph retrieval, demonstrating generalizability to existing graph retrieval and hybrid retrieval methods. Finally, we present a context-aware generation strategy that utilizes the retrieved graph-structured information to generate precise and contextually enriched responses using large language models (LLMs). Extensive experiments on research question answering benchmarks across multiple domains demonstrate that our CG-RAG framework significantly outperforms RAG methods combined with various state-of-the-art retrieval approaches, delivering superior retrieval accuracy and generation quality.
Eliza: A Web3 friendly AI Agent Operating System
Jan 12, 2025Abstract:AI Agent, powered by large language models (LLMs) as its cognitive core, is an intelligent agentic system capable of autonomously controlling and determining the execution paths under user's instructions. With the burst of capabilities of LLMs and various plugins, such as RAG, text-to-image/video/3D, etc., the potential of AI Agents has been vastly expanded, with their capabilities growing stronger by the day. However, at the intersection between AI and web3, there is currently no ideal agentic framework that can seamlessly integrate web3 applications into AI agent functionalities. In this paper, we propose Eliza, the first open-source web3-friendly Agentic framework that makes the deployment of web3 applications effortless. We emphasize that every aspect of Eliza is a regular Typescript program under the full control of its user, and it seamlessly integrates with web3 (i.e., reading and writing blockchain data, interacting with smart contracts, etc.). Furthermore, we show how stable performance is achieved through the pragmatic implementation of the key components of Eliza's runtime. Our code is publicly available at https://github.com/ai16z/eliza.
Representation Learning of Geometric Trees
Aug 16, 2024Abstract:Geometric trees are characterized by their tree-structured layout and spatially constrained nodes and edges, which significantly impacts their topological attributes. This inherent hierarchical structure plays a crucial role in domains such as neuron morphology and river geomorphology, but traditional graph representation methods often overlook these specific characteristics of tree structures. To address this, we introduce a new representation learning framework tailored for geometric trees. It first features a unique message passing neural network, which is both provably geometrical structure-recoverable and rotation-translation invariant. To address the data label scarcity issue, our approach also includes two innovative training targets that reflect the hierarchical ordering and geometric structure of these geometric trees. This enables fully self-supervised learning without explicit labels. We validate our method's effectiveness on eight real-world datasets, demonstrating its capability to represent geometric trees.
GRASP: GRAph-Structured Pyramidal Whole Slide Image Representation
Feb 06, 2024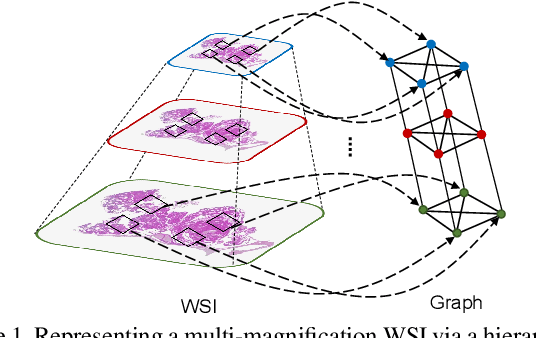
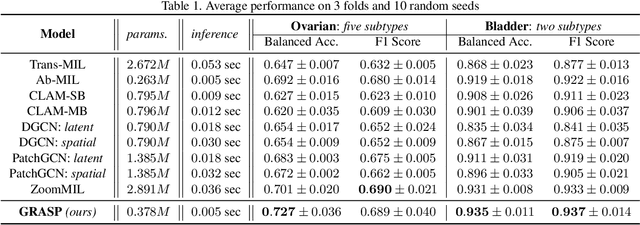
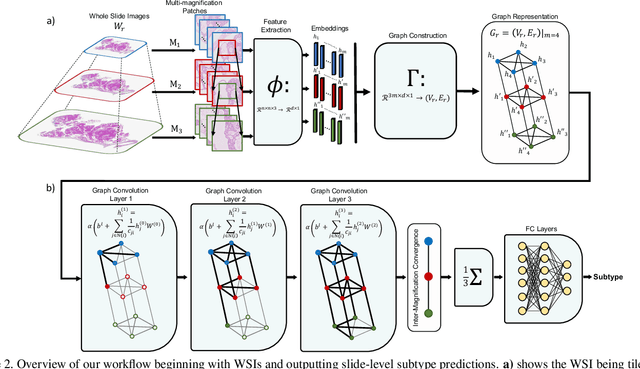
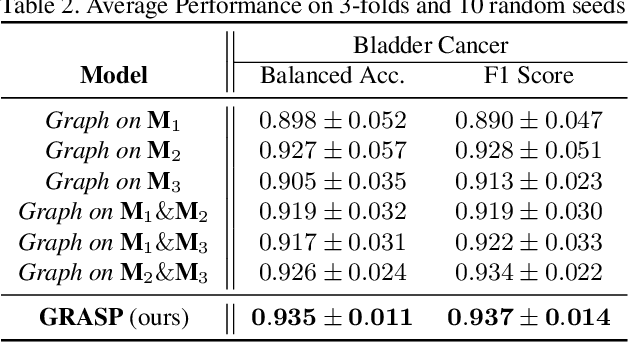
Abstract:Cancer subtyping is one of the most challenging tasks in digital pathology, where Multiple Instance Learning (MIL) by processing gigapixel whole slide images (WSIs) has been in the spotlight of recent research. However, MIL approaches do not take advantage of inter- and intra-magnification information contained in WSIs. In this work, we present GRASP, a novel graph-structured multi-magnification framework for processing WSIs in digital pathology. Our approach is designed to dynamically emulate the pathologist's behavior in handling WSIs and benefits from the hierarchical structure of WSIs. GRASP, which introduces a convergence-based node aggregation instead of traditional pooling mechanisms, outperforms state-of-the-art methods over two distinct cancer datasets by a margin of up to 10% balanced accuracy, while being 7 times smaller than the closest-performing state-of-the-art model in terms of the number of parameters. Our results show that GRASP is dynamic in finding and consulting with different magnifications for subtyping cancers and is reliable and stable across different hyperparameters. The model's behavior has been evaluated by two expert pathologists confirming the interpretability of the model's dynamic. We also provide a theoretical foundation, along with empirical evidence, for our work, explaining how GRASP interacts with different magnifications and nodes in the graph to make predictions. We believe that the strong characteristics yet simple structure of GRASP will encourage the development of interpretable, structure-based designs for WSI representation in digital pathology. Furthermore, we publish two large graph datasets of rare Ovarian and Bladder cancers to contribute to the field.
Non-Euclidean Spatial Graph Neural Network
Dec 17, 2023



Abstract:Spatial networks are networks whose graph topology is constrained by their embedded spatial space. Understanding the coupled spatial-graph properties is crucial for extracting powerful representations from spatial networks. Therefore, merely combining individual spatial and network representations cannot reveal the underlying interaction mechanism of spatial networks. Besides, existing spatial network representation learning methods can only consider networks embedded in Euclidean space, and can not well exploit the rich geometric information carried by irregular and non-uniform non-Euclidean space. In order to address this issue, in this paper we propose a novel generic framework to learn the representation of spatial networks that are embedded in non-Euclidean manifold space. Specifically, a novel message-passing-based neural network is proposed to combine graph topology and spatial geometry, where spatial geometry is extracted as messages on the edges. We theoretically guarantee that the learned representations are provably invariant to important symmetries such as rotation or translation, and simultaneously maintain sufficient ability in distinguishing different geometric structures. The strength of our proposed method is demonstrated through extensive experiments on both synthetic and real-world datasets.
VOLTA: an Environment-Aware Contrastive Cell Representation Learning for Histopathology
Mar 08, 2023
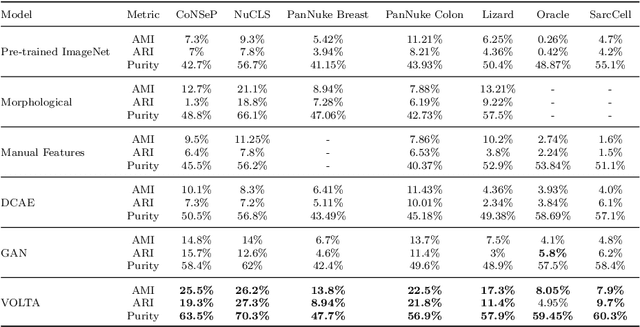

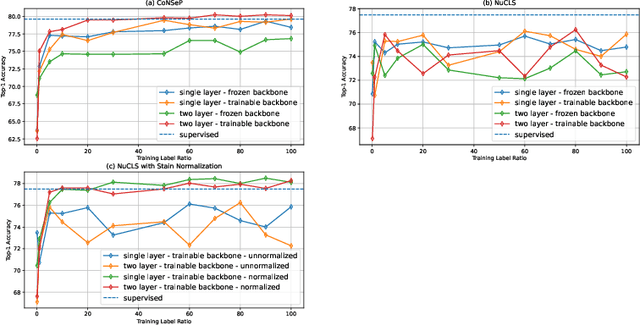
Abstract:In clinical practice, many diagnosis tasks rely on the identification of cells in histopathology images. While supervised machine learning techniques require labels, providing manual cell annotations is time-consuming due to the large number of cells. In this paper, we propose a self-supervised framework (VOLTA) for cell representation learning in histopathology images using a novel technique that accounts for the cell's mutual relationship with its environment for improved cell representations. We subjected our model to extensive experiments on the data collected from multiple institutions around the world comprising of over 700,000 cells, four cancer types, and cell types ranging from three to six categories for each dataset. The results show that our model outperforms the state-of-the-art models in cell representation learning. To showcase the potential power of our proposed framework, we applied VOLTA to ovarian and endometrial cancers with very small sample sizes (10-20 samples) and demonstrated that our cell representations can be utilized to identify the known histotypes of ovarian cancer and provide novel insights that link histopathology and molecular subtypes of endometrial cancer. Unlike supervised deep learning models that require large sample sizes for training, we provide a framework that can empower new discoveries without any annotation data in situations where sample sizes are limited.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge