Ramin Nakhli
HexaGen3D: StableDiffusion is just one step away from Fast and Diverse Text-to-3D Generation
Jan 15, 2024Abstract:Despite the latest remarkable advances in generative modeling, efficient generation of high-quality 3D assets from textual prompts remains a difficult task. A key challenge lies in data scarcity: the most extensive 3D datasets encompass merely millions of assets, while their 2D counterparts contain billions of text-image pairs. To address this, we propose a novel approach which harnesses the power of large, pretrained 2D diffusion models. More specifically, our approach, HexaGen3D, fine-tunes a pretrained text-to-image model to jointly predict 6 orthographic projections and the corresponding latent triplane. We then decode these latents to generate a textured mesh. HexaGen3D does not require per-sample optimization, and can infer high-quality and diverse objects from textual prompts in 7 seconds, offering significantly better quality-to-latency trade-offs when comparing to existing approaches. Furthermore, HexaGen3D demonstrates strong generalization to new objects or compositions.
VOLTA: an Environment-Aware Contrastive Cell Representation Learning for Histopathology
Mar 08, 2023
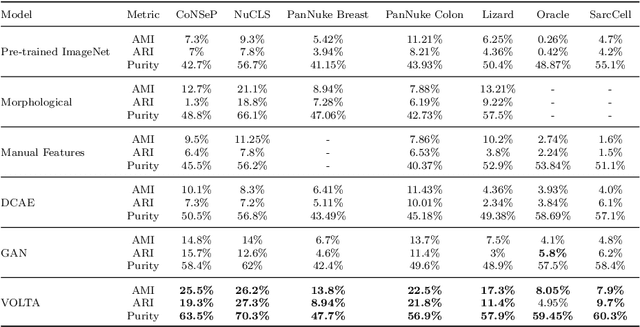

Abstract:In clinical practice, many diagnosis tasks rely on the identification of cells in histopathology images. While supervised machine learning techniques require labels, providing manual cell annotations is time-consuming due to the large number of cells. In this paper, we propose a self-supervised framework (VOLTA) for cell representation learning in histopathology images using a novel technique that accounts for the cell's mutual relationship with its environment for improved cell representations. We subjected our model to extensive experiments on the data collected from multiple institutions around the world comprising of over 700,000 cells, four cancer types, and cell types ranging from three to six categories for each dataset. The results show that our model outperforms the state-of-the-art models in cell representation learning. To showcase the potential power of our proposed framework, we applied VOLTA to ovarian and endometrial cancers with very small sample sizes (10-20 samples) and demonstrated that our cell representations can be utilized to identify the known histotypes of ovarian cancer and provide novel insights that link histopathology and molecular subtypes of endometrial cancer. Unlike supervised deep learning models that require large sample sizes for training, we provide a framework that can empower new discoveries without any annotation data in situations where sample sizes are limited.
AMIGO: Sparse Multi-Modal Graph Transformer with Shared-Context Processing for Representation Learning of Giga-pixel Images
Mar 01, 2023



Abstract:Processing giga-pixel whole slide histopathology images (WSI) is a computationally expensive task. Multiple instance learning (MIL) has become the conventional approach to process WSIs, in which these images are split into smaller patches for further processing. However, MIL-based techniques ignore explicit information about the individual cells within a patch. In this paper, by defining the novel concept of shared-context processing, we designed a multi-modal Graph Transformer (AMIGO) that uses the celluar graph within the tissue to provide a single representation for a patient while taking advantage of the hierarchical structure of the tissue, enabling a dynamic focus between cell-level and tissue-level information. We benchmarked the performance of our model against multiple state-of-the-art methods in survival prediction and showed that ours can significantly outperform all of them including hierarchical Vision Transformer (ViT). More importantly, we show that our model is strongly robust to missing information to an extent that it can achieve the same performance with as low as 20% of the data. Finally, in two different cancer datasets, we demonstrated that our model was able to stratify the patients into low-risk and high-risk groups while other state-of-the-art methods failed to achieve this goal. We also publish a large dataset of immunohistochemistry images (InUIT) containing 1,600 tissue microarray (TMA) cores from 188 patients along with their survival information, making it one of the largest publicly available datasets in this context.
CCRL: Contrastive Cell Representation Learning
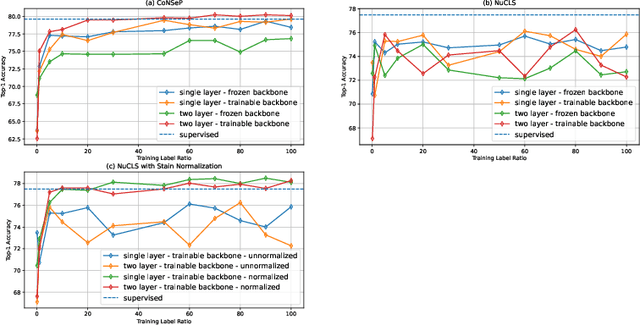
Aug 12, 2022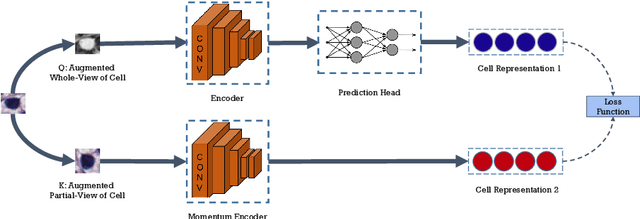
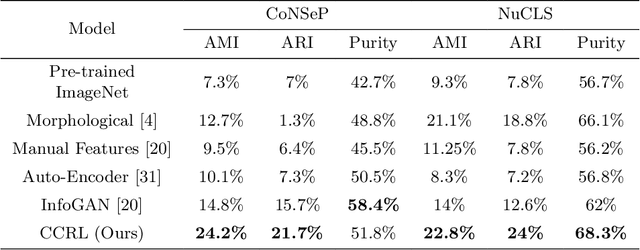
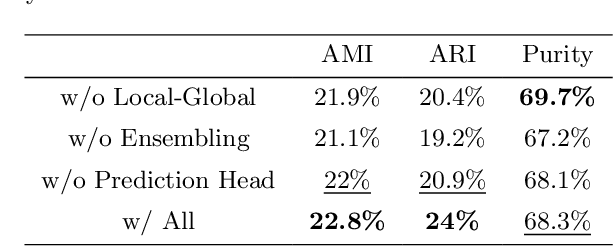
Abstract:Cell identification within the H&E slides is an essential prerequisite that can pave the way towards further pathology analyses including tissue classification, cancer grading, and phenotype prediction. However, performing such a task using deep learning techniques requires a large cell-level annotated dataset. Although previous studies have investigated the performance of contrastive self-supervised methods in tissue classification, the utility of this class of algorithms in cell identification and clustering is still unknown. In this work, we investigated the utility of Self-Supervised Learning (SSL) in cell clustering by proposing the Contrastive Cell Representation Learning (CCRL) model. Through comprehensive comparisons, we show that this model can outperform all currently available cell clustering models by a large margin across two datasets from different tissue types. More interestingly, the results show that our proposed model worked well with a few number of cell categories while the utility of SSL models has been mainly shown in the context of natural image datasets with large numbers of classes (e.g., ImageNet). The unsupervised representation learning approach proposed in this research eliminates the time-consuming step of data annotation in cell classification tasks, which enables us to train our model on a much larger dataset compared to previous methods. Therefore, considering the promising outcome, this approach can open a new avenue to automatic cell representation learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge