CCRL: Contrastive Cell Representation Learning
Paper and Code
Aug 12, 2022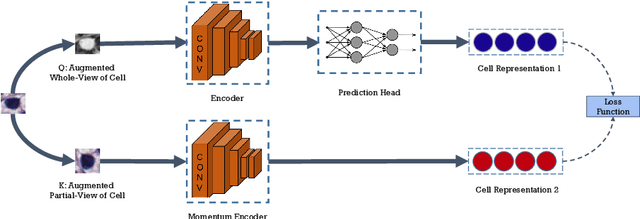
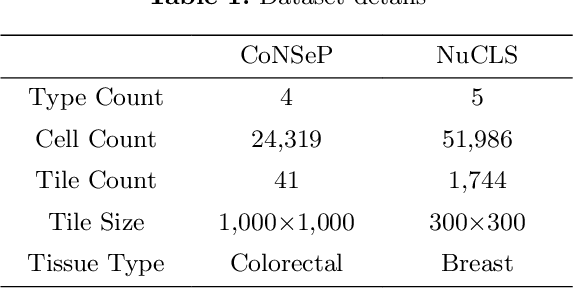
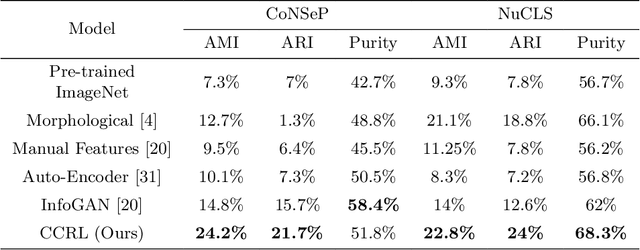
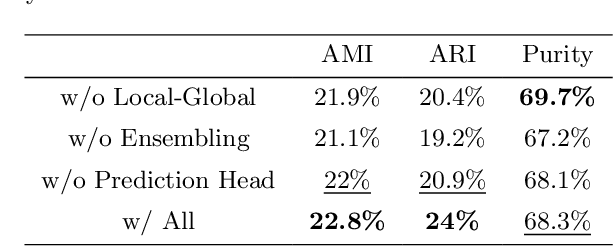
Cell identification within the H&E slides is an essential prerequisite that can pave the way towards further pathology analyses including tissue classification, cancer grading, and phenotype prediction. However, performing such a task using deep learning techniques requires a large cell-level annotated dataset. Although previous studies have investigated the performance of contrastive self-supervised methods in tissue classification, the utility of this class of algorithms in cell identification and clustering is still unknown. In this work, we investigated the utility of Self-Supervised Learning (SSL) in cell clustering by proposing the Contrastive Cell Representation Learning (CCRL) model. Through comprehensive comparisons, we show that this model can outperform all currently available cell clustering models by a large margin across two datasets from different tissue types. More interestingly, the results show that our proposed model worked well with a few number of cell categories while the utility of SSL models has been mainly shown in the context of natural image datasets with large numbers of classes (e.g., ImageNet). The unsupervised representation learning approach proposed in this research eliminates the time-consuming step of data annotation in cell classification tasks, which enables us to train our model on a much larger dataset compared to previous methods. Therefore, considering the promising outcome, this approach can open a new avenue to automatic cell representation learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge