Ali Khajegili Mirabadi
Michael Pokorny
Humanity's Last Exam
Jan 24, 2025Abstract:Benchmarks are important tools for tracking the rapid advancements in large language model (LLM) capabilities. However, benchmarks are not keeping pace in difficulty: LLMs now achieve over 90\% accuracy on popular benchmarks like MMLU, limiting informed measurement of state-of-the-art LLM capabilities. In response, we introduce Humanity's Last Exam (HLE), a multi-modal benchmark at the frontier of human knowledge, designed to be the final closed-ended academic benchmark of its kind with broad subject coverage. HLE consists of 3,000 questions across dozens of subjects, including mathematics, humanities, and the natural sciences. HLE is developed globally by subject-matter experts and consists of multiple-choice and short-answer questions suitable for automated grading. Each question has a known solution that is unambiguous and easily verifiable, but cannot be quickly answered via internet retrieval. State-of-the-art LLMs demonstrate low accuracy and calibration on HLE, highlighting a significant gap between current LLM capabilities and the expert human frontier on closed-ended academic questions. To inform research and policymaking upon a clear understanding of model capabilities, we publicly release HLE at https://lastexam.ai.
Benchmarking Histopathology Foundation Models for Ovarian Cancer Bevacizumab Treatment Response Prediction from Whole Slide Images
Jul 30, 2024



Abstract:Bevacizumab is a widely studied targeted therapeutic drug used in conjunction with standard chemotherapy for the treatment of recurrent ovarian cancer. While its administration has shown to increase the progression-free survival (PFS) in patients with advanced stage ovarian cancer, the lack of identifiable biomarkers for predicting patient response has been a major roadblock in its effective adoption towards personalized medicine. In this work, we leverage the latest histopathology foundation models trained on large-scale whole slide image (WSI) datasets to extract ovarian tumor tissue features for predicting bevacizumab response from WSIs. Our extensive experiments across a combination of different histopathology foundation models and multiple instance learning (MIL) strategies demonstrate capability of these large models in predicting bevacizumab response in ovarian cancer patients with the best models achieving an AUC score of 0.86 and an accuracy score of 72.5%. Furthermore, our survival models are able to stratify high- and low-risk cases with statistical significance (p < 0.05) even among the patients with the aggressive subtype of high-grade serous ovarian carcinoma. This work highlights the utility of histopathology foundation models for the task of ovarian bevacizumab response prediction from WSIs. The high-attention regions of the WSIs highlighted by these models not only aid the model explainability but also serve as promising imaging biomarkers for treatment prognosis.
GRASP: GRAph-Structured Pyramidal Whole Slide Image Representation
Feb 06, 2024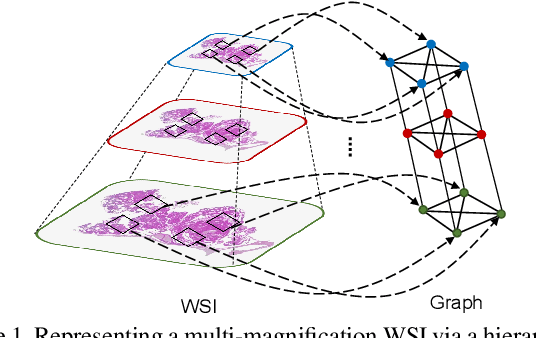
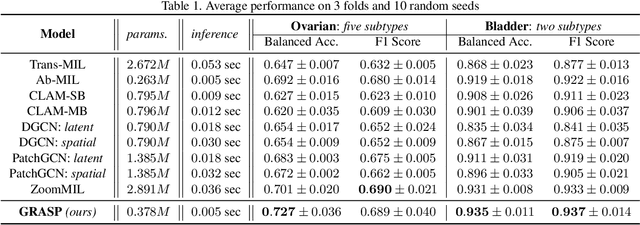
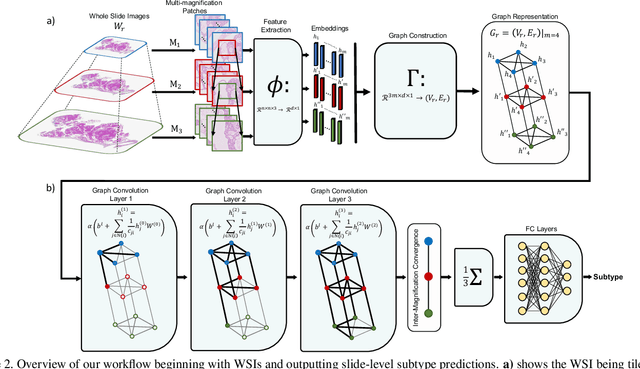
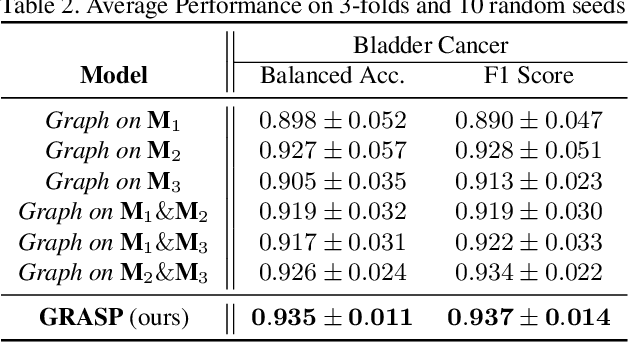
Abstract:Cancer subtyping is one of the most challenging tasks in digital pathology, where Multiple Instance Learning (MIL) by processing gigapixel whole slide images (WSIs) has been in the spotlight of recent research. However, MIL approaches do not take advantage of inter- and intra-magnification information contained in WSIs. In this work, we present GRASP, a novel graph-structured multi-magnification framework for processing WSIs in digital pathology. Our approach is designed to dynamically emulate the pathologist's behavior in handling WSIs and benefits from the hierarchical structure of WSIs. GRASP, which introduces a convergence-based node aggregation instead of traditional pooling mechanisms, outperforms state-of-the-art methods over two distinct cancer datasets by a margin of up to 10% balanced accuracy, while being 7 times smaller than the closest-performing state-of-the-art model in terms of the number of parameters. Our results show that GRASP is dynamic in finding and consulting with different magnifications for subtyping cancers and is reliable and stable across different hyperparameters. The model's behavior has been evaluated by two expert pathologists confirming the interpretability of the model's dynamic. We also provide a theoretical foundation, along with empirical evidence, for our work, explaining how GRASP interacts with different magnifications and nodes in the graph to make predictions. We believe that the strong characteristics yet simple structure of GRASP will encourage the development of interpretable, structure-based designs for WSI representation in digital pathology. Furthermore, we publish two large graph datasets of rare Ovarian and Bladder cancers to contribute to the field.
The Information & Mutual Information Ratio for Counting Image Features and Their Matches
May 14, 2020

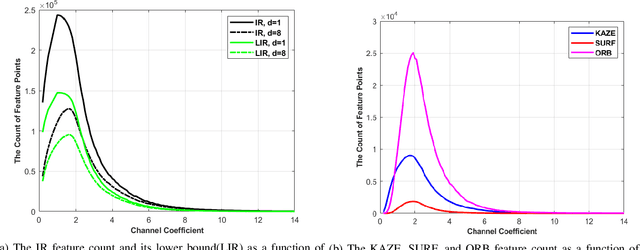
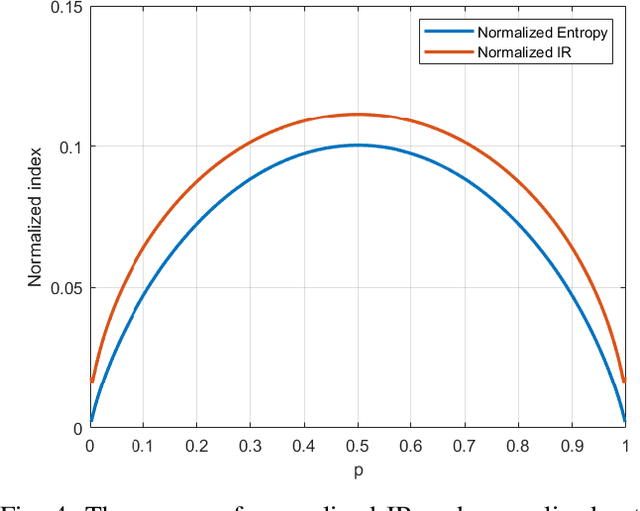
Abstract:Feature extraction and description is an important topic of computer vision, as it is the starting point of a number of tasks such as image reconstruction, stitching, registration, and recognition among many others. In this paper, two new image features are proposed: the Information Ratio (IR) and the Mutual Information Ratio (MIR). The IR is a feature of a single image, while the MIR describes features common across two or more images.We begin by introducing the IR and the MIR and motivate these features in an information theoretical context as the ratio of the self-information of an intensity level over the information contained over the pixels of the same intensity. Notably, the relationship of the IR and MIR with the image entropy and mutual information, classic information measures, are discussed. Finally, the effectiveness of these features is tested through feature extraction over INRIA Copydays datasets and feature matching over the Oxfords Affine Covariant Regions. These numerical evaluations validate the relevance of the IR and MIR in practical computer vision tasks
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge