Mayur Mallya
Benchmarking Histopathology Foundation Models for Ovarian Cancer Bevacizumab Treatment Response Prediction from Whole Slide Images
Jul 30, 2024



Abstract:Bevacizumab is a widely studied targeted therapeutic drug used in conjunction with standard chemotherapy for the treatment of recurrent ovarian cancer. While its administration has shown to increase the progression-free survival (PFS) in patients with advanced stage ovarian cancer, the lack of identifiable biomarkers for predicting patient response has been a major roadblock in its effective adoption towards personalized medicine. In this work, we leverage the latest histopathology foundation models trained on large-scale whole slide image (WSI) datasets to extract ovarian tumor tissue features for predicting bevacizumab response from WSIs. Our extensive experiments across a combination of different histopathology foundation models and multiple instance learning (MIL) strategies demonstrate capability of these large models in predicting bevacizumab response in ovarian cancer patients with the best models achieving an AUC score of 0.86 and an accuracy score of 72.5%. Furthermore, our survival models are able to stratify high- and low-risk cases with statistical significance (p < 0.05) even among the patients with the aggressive subtype of high-grade serous ovarian carcinoma. This work highlights the utility of histopathology foundation models for the task of ovarian bevacizumab response prediction from WSIs. The high-attention regions of the WSIs highlighted by these models not only aid the model explainability but also serve as promising imaging biomarkers for treatment prognosis.
Deep Multimodal Guidance for Medical Image Classification
Mar 10, 2022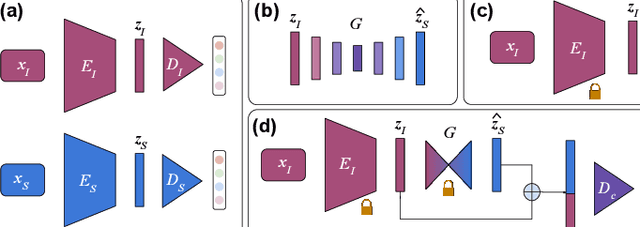
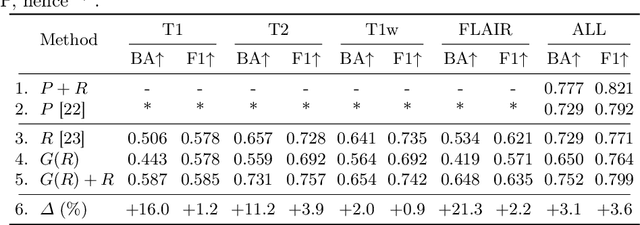
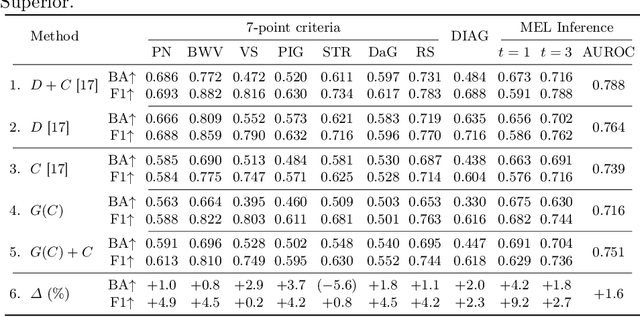
Abstract:Medical imaging is a cornerstone of therapy and diagnosis in modern medicine. However, the choice of imaging modality for a particular theranostic task typically involves trade-offs between the feasibility of using a particular modality (e.g., short wait times, low cost, fast acquisition, reduced radiation/invasiveness) and the expected performance on a clinical task (e.g., diagnostic accuracy, efficacy of treatment planning and guidance). In this work, we aim to apply the knowledge learned from the less feasible but better-performing (superior) modality to guide the utilization of the more-feasible yet under-performing (inferior) modality and steer it towards improved performance. We focus on the application of deep learning for image-based diagnosis. We develop a light-weight guidance model that leverages the latent representation learned from the superior modality, when training a model that consumes only the inferior modality. We examine the advantages of our method in the context of two clinical applications: multi-task skin lesion classification from clinical and dermoscopic images and brain tumor classification from multi-sequence magnetic resonance imaging (MRI) and histopathology images. For both these scenarios we show a boost in diagnostic performance of the inferior modality without requiring the superior modality. Furthermore, in the case of brain tumor classification, our method outperforms the model trained on the superior modality while producing comparable results to the model that uses both modalities during inference.
Applying Artificial Intelligence to Glioma Imaging: Advances and Challenges
Nov 28, 2019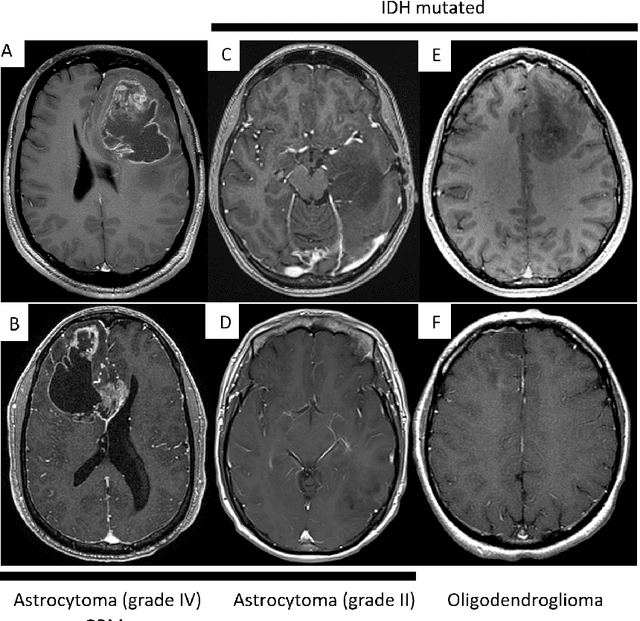
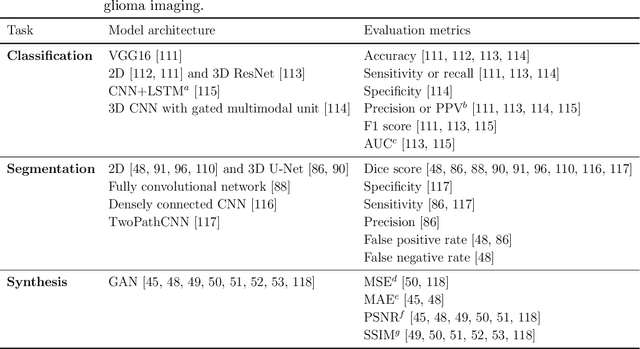
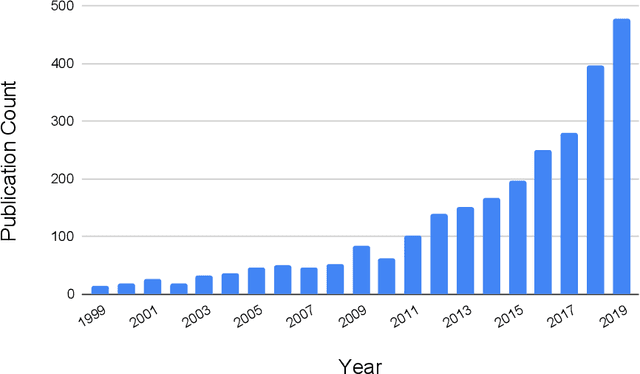
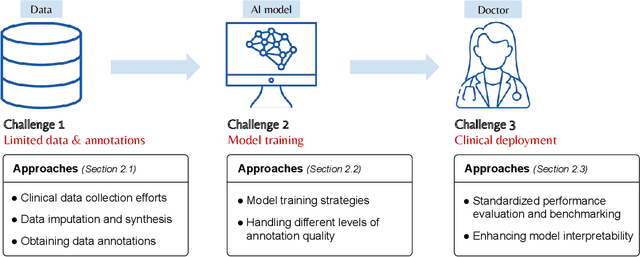
Abstract:Primary brain tumors including gliomas continue to pose significant management challenges to clinicians. While the presentation, the pathology, and the clinical course of these lesions is variable, the initial investigations are usually similar. Patients who are suspected to have a brain tumor will be assessed with computed tomography (CT) and magnetic resonance imaging (MRI). The imaging findings are used by neurosurgeons to determine the feasibility of surgical resection and plan such an undertaking. Imaging studies are also an indispensable tool in tracking tumor progression or its response to treatment. As these imaging studies are non-invasive, relatively cheap and accessible to patients, there have been many efforts over the past two decades to increase the amount of clinically-relevant information that can be extracted from brain imaging. Most recently, artificial intelligence (AI) techniques have been employed to segment and characterize brain tumors, as well as to detect progression or treatment-response. However, the clinical utility of such endeavours remains limited due to challenges in data collection and annotation, model training, and in the reliability of AI-generated information. We provide a review of recent advances in addressing the above challenges. First, to overcome the challenge of data paucity, different image imputation and synthesis techniques along with annotation collection efforts are summarized. Next, various training strategies are presented to meet multiple desiderata, such as model performance, generalization ability, data privacy protection, and learning with sparse annotations. Finally, standardized performance evaluation and model interpretability methods have been reviewed. We believe that these technical approaches will facilitate the development of a fully-functional AI tool in the clinical care of patients with gliomas.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge