C. Blake Gilks
GRASP: GRAph-Structured Pyramidal Whole Slide Image Representation
Feb 06, 2024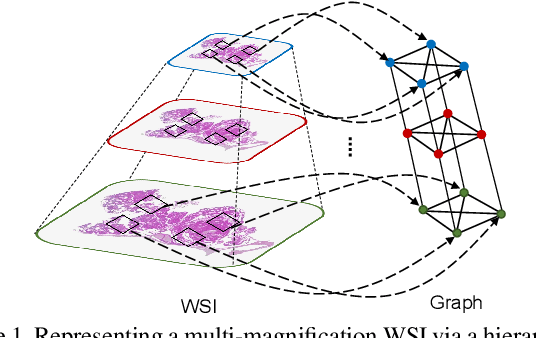
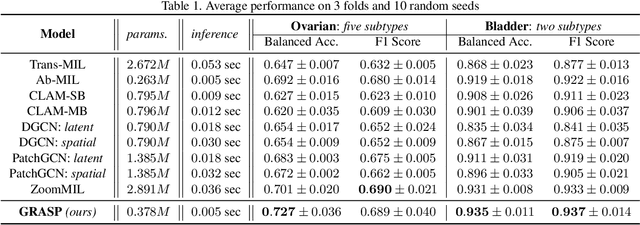
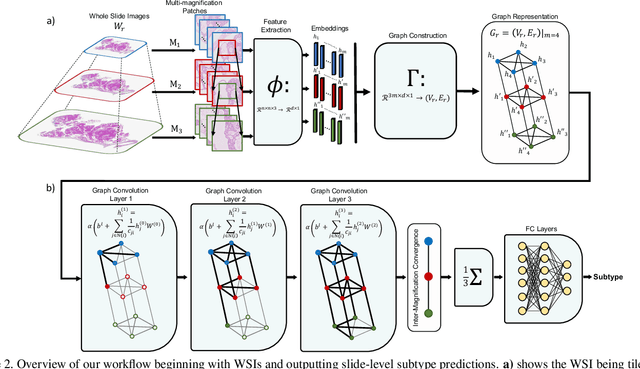
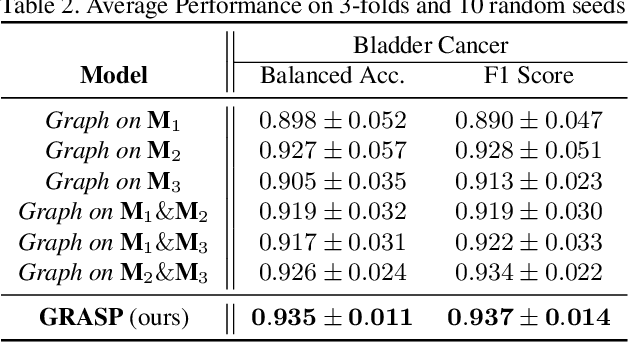
Abstract:Cancer subtyping is one of the most challenging tasks in digital pathology, where Multiple Instance Learning (MIL) by processing gigapixel whole slide images (WSIs) has been in the spotlight of recent research. However, MIL approaches do not take advantage of inter- and intra-magnification information contained in WSIs. In this work, we present GRASP, a novel graph-structured multi-magnification framework for processing WSIs in digital pathology. Our approach is designed to dynamically emulate the pathologist's behavior in handling WSIs and benefits from the hierarchical structure of WSIs. GRASP, which introduces a convergence-based node aggregation instead of traditional pooling mechanisms, outperforms state-of-the-art methods over two distinct cancer datasets by a margin of up to 10% balanced accuracy, while being 7 times smaller than the closest-performing state-of-the-art model in terms of the number of parameters. Our results show that GRASP is dynamic in finding and consulting with different magnifications for subtyping cancers and is reliable and stable across different hyperparameters. The model's behavior has been evaluated by two expert pathologists confirming the interpretability of the model's dynamic. We also provide a theoretical foundation, along with empirical evidence, for our work, explaining how GRASP interacts with different magnifications and nodes in the graph to make predictions. We believe that the strong characteristics yet simple structure of GRASP will encourage the development of interpretable, structure-based designs for WSI representation in digital pathology. Furthermore, we publish two large graph datasets of rare Ovarian and Bladder cancers to contribute to the field.
Classification of Epithelial Ovarian Carcinoma Whole-Slide Pathology Images Using Deep Transfer Learning
May 22, 2020Abstract:Ovarian cancer is the most lethal cancer of the female reproductive organs. There are $5$ major histological subtypes of epithelial ovarian cancer, each with distinct morphological, genetic, and clinical features. Currently, these histotypes are determined by a pathologist's microscopic examination of tumor whole-slide images (WSI). This process has been hampered by poor inter-observer agreement (Cohen's kappa $0.54$-$0.67$). We utilized a \textit{two}-stage deep transfer learning algorithm based on convolutional neural networks (CNN) and progressive resizing for automatic classification of epithelial ovarian carcinoma WSIs. The proposed algorithm achieved a mean accuracy of $87.54\%$ and Cohen's kappa of $0.8106$ in the slide-level classification of $305$ WSIs; performing better than a standard CNN and pathologists without gynecology-specific training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge