Giorgio Ascoli
Representation Learning of Geometric Trees
Aug 16, 2024Abstract:Geometric trees are characterized by their tree-structured layout and spatially constrained nodes and edges, which significantly impacts their topological attributes. This inherent hierarchical structure plays a crucial role in domains such as neuron morphology and river geomorphology, but traditional graph representation methods often overlook these specific characteristics of tree structures. To address this, we introduce a new representation learning framework tailored for geometric trees. It first features a unique message passing neural network, which is both provably geometrical structure-recoverable and rotation-translation invariant. To address the data label scarcity issue, our approach also includes two innovative training targets that reflect the hierarchical ordering and geometric structure of these geometric trees. This enables fully self-supervised learning without explicit labels. We validate our method's effectiveness on eight real-world datasets, demonstrating its capability to represent geometric trees.
BEAN: Interpretable Representation Learning with Biologically-Enhanced Artificial Neuronal Assembly Regularization
Sep 27, 2019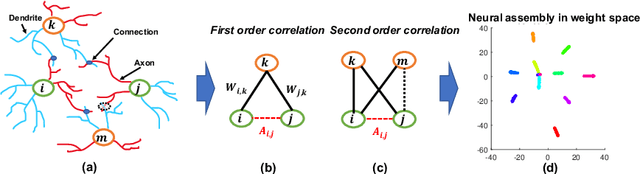
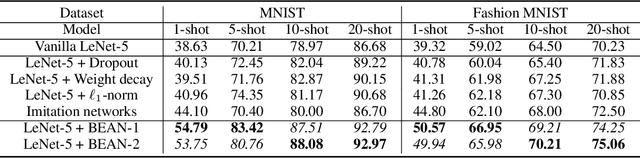
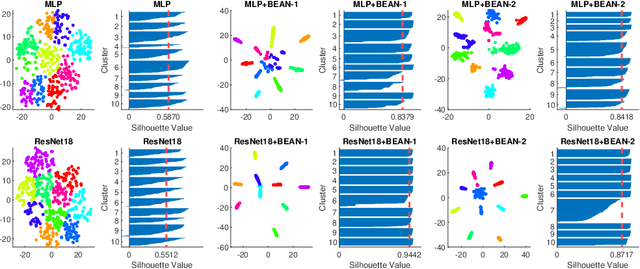
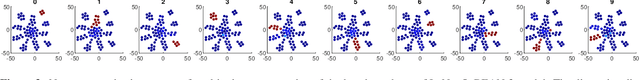
Abstract:Deep neural networks (DNNs) are known for extracting good representations from a large amount of data. However, the representations learned in DNNs are typically hard to interpret, especially the ones learned in dense layers. One crucial issue is that neurons within each layer of DNNs are conditionally independent with each other, which makes the co-training and analysis of neurons at higher modularity difficult. In contrast, the dependency patterns of biological neurons in the human brain are largely different from those of DNNs. Neuronal assembly describes such neuron dependencies that could be found among a group of biological neurons as having strong internal synaptic interactions, potentially high semantical correlations that are deemed to facilitate the memorization process. In this paper, we show such a crucial gap between DNNs and biological neural networks (BNNs) can be bridged by the newly proposed Biologically-Enhanced Artificial Neuronal assembly (BEAN) regularization that could enforce dependencies among neurons in dense layers of DNNs without altering the conventional architecture. Both qualitative and quantitative analyses show that BEAN enables the formations of interpretable and biologically plausible neuronal assemblies in dense layers and consequently enhances the modularity and interpretability of the hidden representations learned. Moreover, BEAN further results in sparse and structured connectivity and parameter sharing among neurons, which substantially improves the efficiency and generalizability of the model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge