Alison Q O'Neil
Controllable Chest X-Ray Report Generation from Longitudinal Representations
Oct 09, 2023



Abstract:Radiology reports are detailed text descriptions of the content of medical scans. Each report describes the presence/absence and location of relevant clinical findings, commonly including comparison with prior exams of the same patient to describe how they evolved. Radiology reporting is a time-consuming process, and scan results are often subject to delays. One strategy to speed up reporting is to integrate automated reporting systems, however clinical deployment requires high accuracy and interpretability. Previous approaches to automated radiology reporting generally do not provide the prior study as input, precluding comparison which is required for clinical accuracy in some types of scans, and offer only unreliable methods of interpretability. Therefore, leveraging an existing visual input format of anatomical tokens, we introduce two novel aspects: (1) longitudinal representation learning -- we input the prior scan as an additional input, proposing a method to align, concatenate and fuse the current and prior visual information into a joint longitudinal representation which can be provided to the multimodal report generation model; (2) sentence-anatomy dropout -- a training strategy for controllability in which the report generator model is trained to predict only sentences from the original report which correspond to the subset of anatomical regions given as input. We show through in-depth experiments on the MIMIC-CXR dataset how the proposed approach achieves state-of-the-art results while enabling anatomy-wise controllable report generation.
Diffusion Models for Causal Discovery via Topological Ordering
Oct 12, 2022



Abstract:Discovering causal relations from observational data becomes possible with additional assumptions such as considering the functional relations to be constrained as nonlinear with additive noise. In this case, the Hessian of the data log-likelihood can be used for finding leaf nodes in a causal graph. Topological ordering approaches for causal discovery exploit this by performing graph discovery in two steps, first sequentially identifying nodes in reverse order of depth (topological ordering), and secondly pruning the potential relations. This is more efficient since the search is performed over a permutation rather than a graph space. However, existing computational methods for obtaining the Hessian still do not scale as the number of variables and the number of samples are increased. Therefore, inspired by recent innovations in diffusion probabilistic models (DPMs), we propose DiffAN, a topological ordering algorithm that leverages DPMs. Further, we introduce theory for updating the learned Hessian without re-training the neural network, and we show that computing with a subset of samples gives an accurate approximation of the ordering, which allows scaling to datasets with more samples and variables. We show empirically that our method scales exceptionally well to datasets with up to $500$ nodes and up to $10^5$ samples while still performing on par over small datasets with state-of-the-art causal discovery methods. Implementation is available at https://github.com/vios-s/DiffAN .
Teacher-Student chain for efficient semi-supervised histology image classification
Mar 20, 2020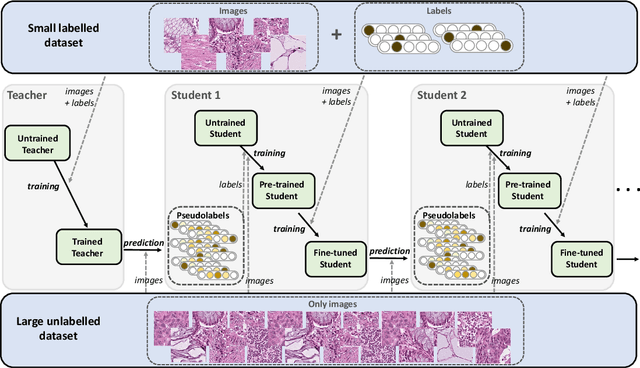

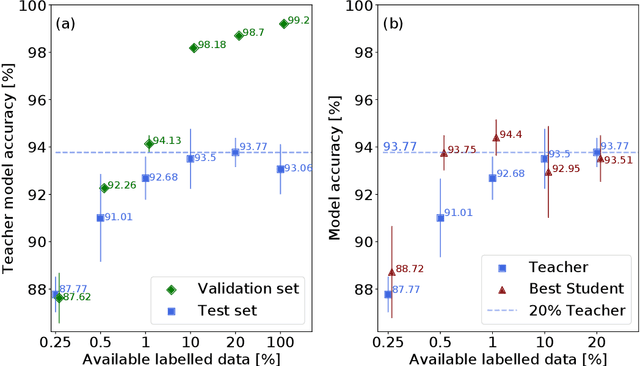
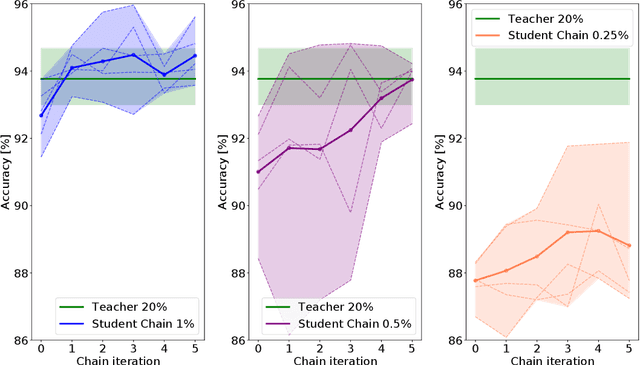
Abstract:Deep learning shows great potential for the domain of digital pathology. An automated digital pathology system could serve as a second reader, perform initial triage in large screening studies, or assist in reporting. However, it is expensive to exhaustively annotate large histology image databases, since medical specialists are a scarce resource. In this paper, we apply the semi-supervised teacher-student knowledge distillation technique proposed by Yalniz et al. (2019) to the task of quantifying prognostic features in colorectal cancer. We obtain accuracy improvements through extending this approach to a chain of students, where each student's predictions are used to train the next student i.e. the student becomes the teacher. Using the chain approach, and only 0.5% labelled data (the remaining 99.5% in the unlabelled pool), we match the accuracy of training on 100% labelled data. At lower percentages of labelled data, similar gains in accuracy are seen, allowing some recovery of accuracy even from a poor initial choice of labelled training set. In conclusion, this approach shows promise for reducing the annotation burden, thus increasing the affordability of automated digital pathology systems.
Language Transfer for Early Warning of Epidemics from Social Media
Oct 10, 2019



Abstract:Statements on social media can be analysed to identify individuals who are experiencing red flag medical symptoms, allowing early detection of the spread of disease such as influenza. Since disease does not respect cultural borders and may spread between populations speaking different languages, we would like to build multilingual models. However, the data required to train models for every language may be difficult, expensive and time-consuming to obtain, particularly for low-resource languages. Taking Japanese as our target language, we explore methods by which data in one language might be used to build models for a different language. We evaluate strategies of training on machine translated data and of zero-shot transfer through the use of multilingual models. We find that the choice of source language impacts the performance, with Chinese-Japanese being a better language pair than English-Japanese. Training on machine translated data shows promise, especially when used in conjunction with a small amount of target language data.
Attaining human-level performance with atlas location autocontext for anatomical landmark detection in 3D CT data
Sep 30, 2018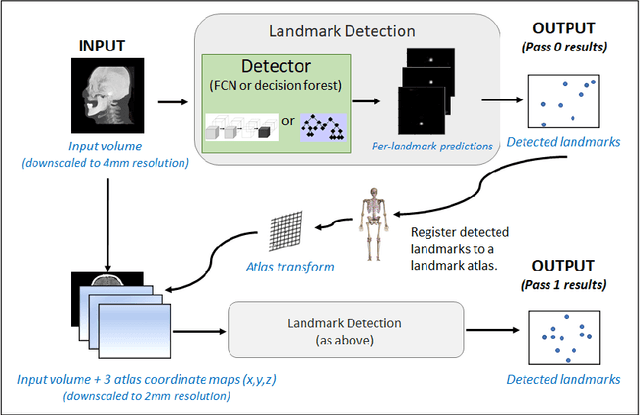
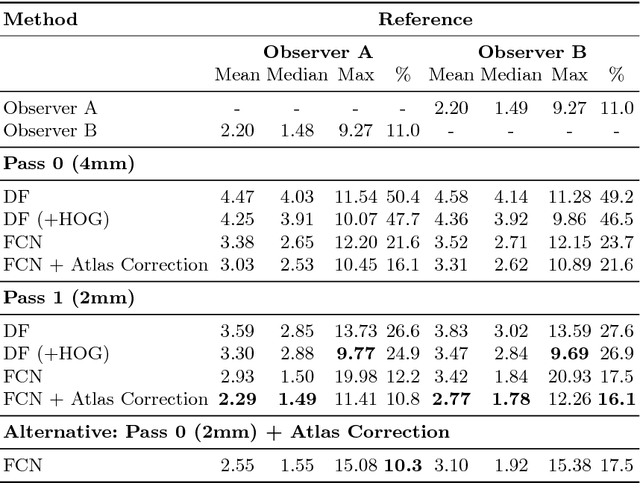

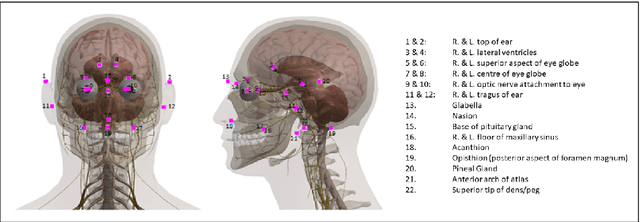
Abstract:We present an efficient neural network method for locating anatomical landmarks in 3D medical CT scans, using atlas location autocontext in order to learn long-range spatial context. Location predictions are made by regression to Gaussian heatmaps, one heatmap per landmark. This system allows patchwise application of a shallow network, thus enabling multiple volumetric heatmaps to be predicted concurrently without prohibitive GPU memory requirements. Further, the system allows inter-landmark spatial relationships to be exploited using a simple overdetermined affine mapping that is robust to detection failures and occlusion or partial views. Evaluation is performed for 22 landmarks defined on a range of structures in head CT scans. Models are trained and validated on 201 scans. Over the final test set of 20 scans which was independently annotated by 2 human annotators, the neural network reaches an accuracy which matches the annotator variability, with similar human and machine patterns of variability across landmark classes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge