Antanas Kascenas
Automated clinical coding using off-the-shelf large language models
Oct 10, 2023Abstract:The task of assigning diagnostic ICD codes to patient hospital admissions is typically performed by expert human coders. Efforts towards automated ICD coding are dominated by supervised deep learning models. However, difficulties in learning to predict the large number of rare codes remain a barrier to adoption in clinical practice. In this work, we leverage off-the-shelf pre-trained generative large language models (LLMs) to develop a practical solution that is suitable for zero-shot and few-shot code assignment. Unsupervised pre-training alone does not guarantee precise knowledge of the ICD ontology and specialist clinical coding task, therefore we frame the task as information extraction, providing a description of each coded concept and asking the model to retrieve related mentions. For efficiency, rather than iterating over all codes, we leverage the hierarchical nature of the ICD ontology to sparsely search for relevant codes. Then, in a second stage, which we term 'meta-refinement', we utilise GPT-4 to select a subset of the relevant labels as predictions. We validate our method using Llama-2, GPT-3.5 and GPT-4 on the CodiEsp dataset of ICD-coded clinical case documents. Our tree-search method achieves state-of-the-art performance on rarer classes, achieving the best macro-F1 of 0.225, whilst achieving slightly lower micro-F1 of 0.157, compared to 0.216 and 0.219 respectively from PLM-ICD. To the best of our knowledge, this is the first method for automated ICD coding requiring no task-specific learning.
Compositional Representation Learning for Brain Tumour Segmentation
Oct 10, 2023Abstract:For brain tumour segmentation, deep learning models can achieve human expert-level performance given a large amount of data and pixel-level annotations. However, the expensive exercise of obtaining pixel-level annotations for large amounts of data is not always feasible, and performance is often heavily reduced in a low-annotated data regime. To tackle this challenge, we adapt a mixed supervision framework, vMFNet, to learn robust compositional representations using unsupervised learning and weak supervision alongside non-exhaustive pixel-level pathology labels. In particular, we use the BraTS dataset to simulate a collection of 2-point expert pathology annotations indicating the top and bottom slice of the tumour (or tumour sub-regions: peritumoural edema, GD-enhancing tumour, and the necrotic / non-enhancing tumour) in each MRI volume, from which weak image-level labels that indicate the presence or absence of the tumour (or the tumour sub-regions) in the image are constructed. Then, vMFNet models the encoded image features with von-Mises-Fisher (vMF) distributions, via learnable and compositional vMF kernels which capture information about structures in the images. We show that good tumour segmentation performance can be achieved with a large amount of weakly labelled data but only a small amount of fully-annotated data. Interestingly, emergent learning of anatomical structures occurs in the compositional representation even given only supervision relating to pathology (tumour).
The role of noise in denoising models for anomaly detection in medical images
Jan 19, 2023



Abstract:Pathological brain lesions exhibit diverse appearance in brain images, in terms of intensity, texture, shape, size, and location. Comprehensive sets of data and annotations are difficult to acquire. Therefore, unsupervised anomaly detection approaches have been proposed using only normal data for training, with the aim of detecting outlier anomalous voxels at test time. Denoising methods, for instance classical denoising autoencoders (DAEs) and more recently emerging diffusion models, are a promising approach, however naive application of pixelwise noise leads to poor anomaly detection performance. We show that optimization of the spatial resolution and magnitude of the noise improves the performance of different model training regimes, with similar noise parameter adjustments giving good performance for both DAEs and diffusion models. Visual inspection of the reconstructions suggests that the training noise influences the trade-off between the extent of the detail that is reconstructed and the extent of erasure of anomalies, both of which contribute to better anomaly detection performance. We validate our findings on two real-world datasets (tumor detection in brain MRI and hemorrhage/ischemia/tumor detection in brain CT), showing good detection on diverse anomaly appearances. Overall, we find that a DAE trained with coarse noise is a fast and simple method that gives state-of-the-art accuracy. Diffusion models applied to anomaly detection are as yet in their infancy and provide a promising avenue for further research.
What is Healthy? Generative Counterfactual Diffusion for Lesion Localization
Jul 25, 2022

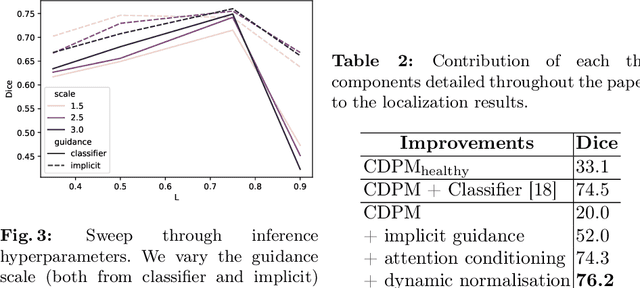

Abstract:Reducing the requirement for densely annotated masks in medical image segmentation is important due to cost constraints. In this paper, we consider the problem of inferring pixel-level predictions of brain lesions by only using image-level labels for training. By leveraging recent advances in generative diffusion probabilistic models (DPM), we synthesize counterfactuals of "How would a patient appear if X pathology was not present?". The difference image between the observed patient state and the healthy counterfactual can be used for inferring the location of pathology. We generate counterfactuals that correspond to the minimal change of the input such that it is transformed to healthy domain. This requires training with healthy and unhealthy data in DPMs. We improve on previous counterfactual DPMs by manipulating the generation process with implicit guidance along with attention conditioning instead of using classifiers. Code is available at https://github.com/vios-s/Diff-SCM.
Attaining human-level performance with atlas location autocontext for anatomical landmark detection in 3D CT data
Sep 30, 2018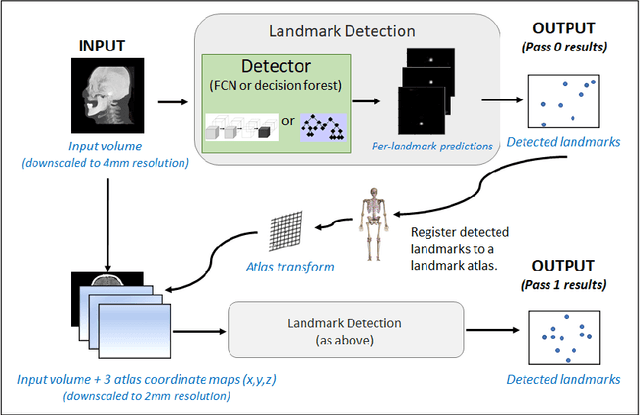
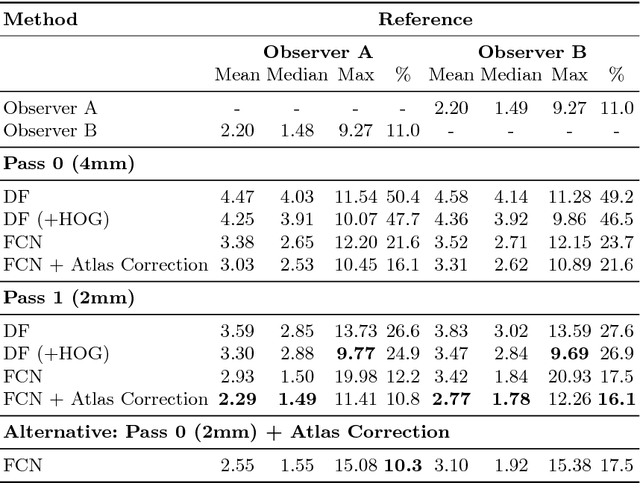

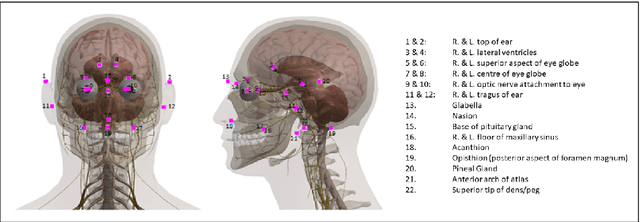
Abstract:We present an efficient neural network method for locating anatomical landmarks in 3D medical CT scans, using atlas location autocontext in order to learn long-range spatial context. Location predictions are made by regression to Gaussian heatmaps, one heatmap per landmark. This system allows patchwise application of a shallow network, thus enabling multiple volumetric heatmaps to be predicted concurrently without prohibitive GPU memory requirements. Further, the system allows inter-landmark spatial relationships to be exploited using a simple overdetermined affine mapping that is robust to detection failures and occlusion or partial views. Evaluation is performed for 22 landmarks defined on a range of structures in head CT scans. Models are trained and validated on 201 scans. Over the final test set of 20 scans which was independently annotated by 2 human annotators, the neural network reaches an accuracy which matches the annotator variability, with similar human and machine patterns of variability across landmark classes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge