Jeremy P. Voisey
The role of noise in denoising models for anomaly detection in medical images
Jan 19, 2023



Abstract:Pathological brain lesions exhibit diverse appearance in brain images, in terms of intensity, texture, shape, size, and location. Comprehensive sets of data and annotations are difficult to acquire. Therefore, unsupervised anomaly detection approaches have been proposed using only normal data for training, with the aim of detecting outlier anomalous voxels at test time. Denoising methods, for instance classical denoising autoencoders (DAEs) and more recently emerging diffusion models, are a promising approach, however naive application of pixelwise noise leads to poor anomaly detection performance. We show that optimization of the spatial resolution and magnitude of the noise improves the performance of different model training regimes, with similar noise parameter adjustments giving good performance for both DAEs and diffusion models. Visual inspection of the reconstructions suggests that the training noise influences the trade-off between the extent of the detail that is reconstructed and the extent of erasure of anomalies, both of which contribute to better anomaly detection performance. We validate our findings on two real-world datasets (tumor detection in brain MRI and hemorrhage/ischemia/tumor detection in brain CT), showing good detection on diverse anomaly appearances. Overall, we find that a DAE trained with coarse noise is a fast and simple method that gives state-of-the-art accuracy. Diffusion models applied to anomaly detection are as yet in their infancy and provide a promising avenue for further research.
Causal Machine Learning for Healthcare and Precision Medicine
May 31, 2022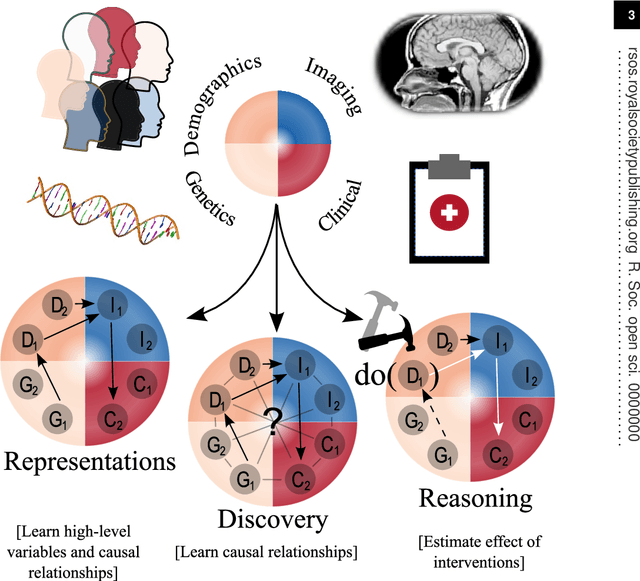
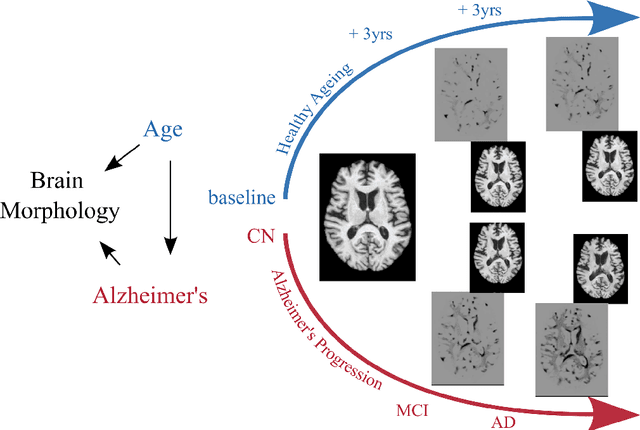
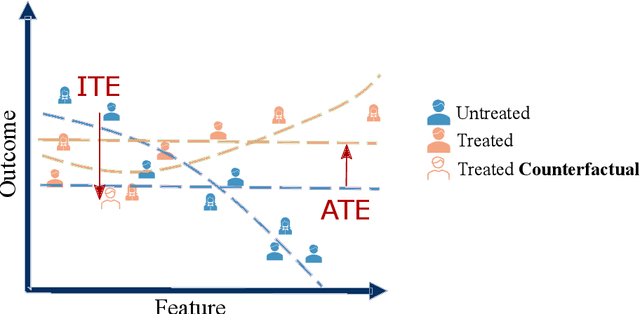
Abstract:Causal machine learning (CML) has experienced increasing popularity in healthcare. Beyond the inherent capabilities of adding domain knowledge into learning systems, CML provides a complete toolset for investigating how a system would react to an intervention (e.g.\ outcome given a treatment). Quantifying effects of interventions allows actionable decisions to be made whilst maintaining robustness in the presence of confounders. Here, we explore how causal inference can be incorporated into different aspects of clinical decision support (CDS) systems by using recent advances in machine learning. Throughout this paper, we use Alzheimer's disease (AD) to create examples for illustrating how CML can be advantageous in clinical scenarios. Furthermore, we discuss important challenges present in healthcare applications such as processing high-dimensional and unstructured data, generalisation to out-of-distribution samples, and temporal relationships, that despite the great effort from the research community remain to be solved. Finally, we review lines of research within causal representation learning, causal discovery and causal reasoning which offer the potential towards addressing the aforementioned challenges.
Pseudo-label refinement using superpixels for semi-supervised brain tumour segmentation
Oct 16, 2021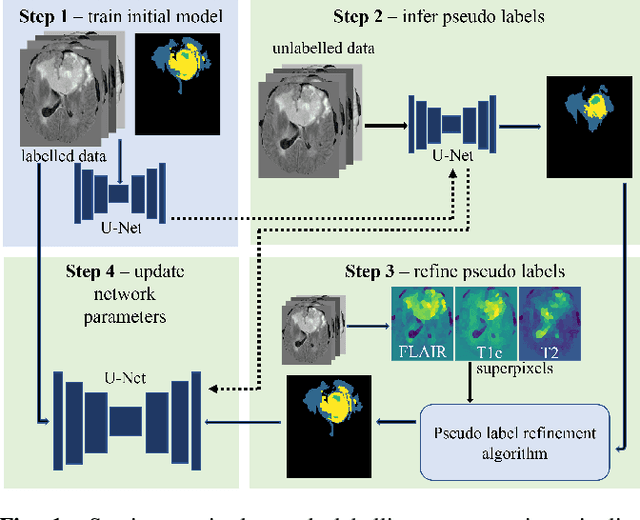
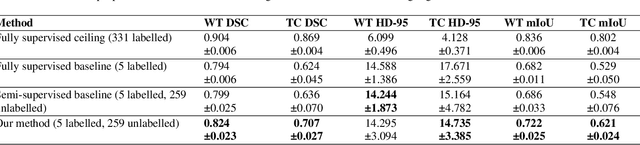
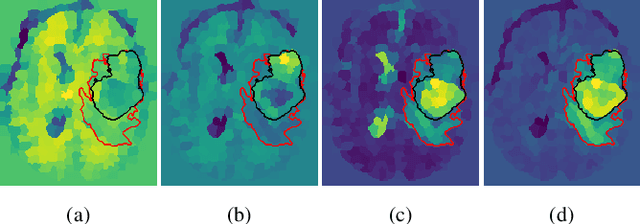
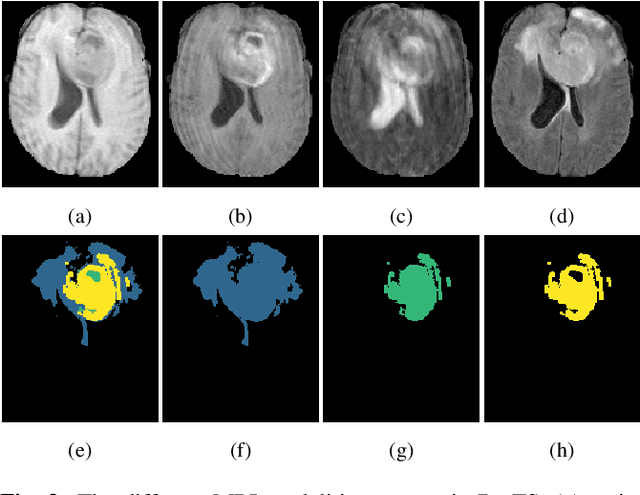
Abstract:Training neural networks using limited annotations is an important problem in the medical domain. Deep Neural Networks (DNNs) typically require large, annotated datasets to achieve acceptable performance which, in the medical domain, are especially difficult to obtain as they require significant time from expert radiologists. Semi-supervised learning aims to overcome this problem by learning segmentations with very little annotated data, whilst exploiting large amounts of unlabelled data. However, the best-known technique, which utilises inferred pseudo-labels, is vulnerable to inaccurate pseudo-labels degrading the performance. We propose a framework based on superpixels - meaningful clusters of adjacent pixels - to improve the accuracy of the pseudo labels and address this issue. Our framework combines superpixels with semi-supervised learning, refining the pseudo-labels during training using the features and edges of the superpixel maps. This method is evaluated on a multimodal magnetic resonance imaging (MRI) dataset for the task of brain tumour region segmentation. Our method demonstrates improved performance over the standard semi-supervised pseudo-labelling baseline when there is a reduced annotator burden and only 5 annotated patients are available. We report DSC=0.824 and DSC=0.707 for the test set whole tumour and tumour core regions respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge