Zhouxin Yu
Scaling Up Dynamic Graph Representation Learning via Spiking Neural Networks
Aug 15, 2022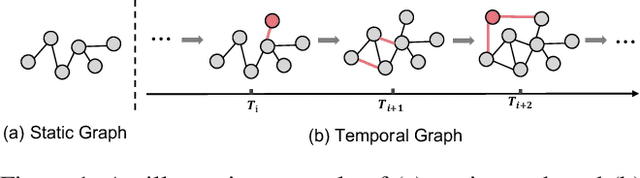



Abstract:Recent years have seen a surge in research on dynamic graph representation learning, which aims to model temporal graphs that are dynamic and evolving constantly over time. However, current work typically models graph dynamics with recurrent neural networks (RNNs), making them suffer seriously from computation and memory overheads on large temporal graphs. So far, scalability of dynamic graph representation learning on large temporal graphs remains one of the major challenges. In this paper, we present a scalable framework, namely SpikeNet, to efficiently capture the temporal and structural patterns of temporal graphs. We explore a new direction in that we can capture the evolving dynamics of temporal graphs with spiking neural networks (SNNs) instead of RNNs. As a low-power alternative to RNNs, SNNs explicitly model graph dynamics as spike trains of neuron populations and enable spike-based propagation in an efficient way. Experiments on three large real-world temporal graph datasets demonstrate that SpikeNet outperforms strong baselines on the temporal node classification task with lower computational costs. Particularly, SpikeNet generalizes to a large temporal graph (2M nodes and 13M edges) with significantly fewer parameters and computation overheads. Our code is publicly available at https://github.com/EdisonLeeeee/SpikeNet
Predicting microRNA-disease associations from knowledge graph using tensor decomposition with relational constraints
Nov 13, 2019



Abstract:Motivation: MiRNAs are a kind of small non-coding RNAs that are not translated into proteins, and aberrant expression of miRNAs is associated with human diseases. Since miRNAs have different roles in diseases, the miRNA-disease associations are categorized into multiple types according to their roles. Predicting miRNA-disease associations and types is critical to understand the underlying pathogenesis of human diseases from the molecular level. Results: In this paper, we formulate the problem as a link prediction in knowledge graphs. We use biomedical knowledge bases to build a knowledge graph of entities representing miRNAs and disease and multi-relations, and we propose a tensor decomposition-based model named TDRC to predict miRNA-disease associations and their types from the knowledge graph. We have experimentally evaluated our method and compared it to several baseline methods. The results demonstrate that the proposed method has high-accuracy and high-efficiency performances.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge