Zhi Jing
HumanoidGen: Data Generation for Bimanual Dexterous Manipulation via LLM Reasoning
Jul 01, 2025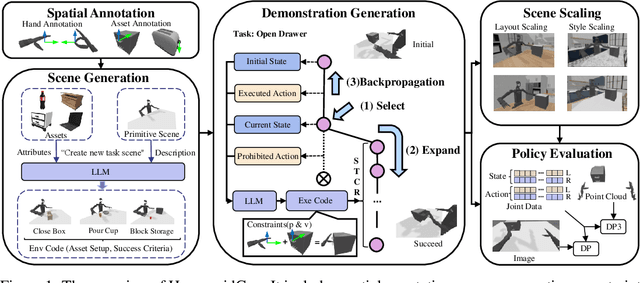
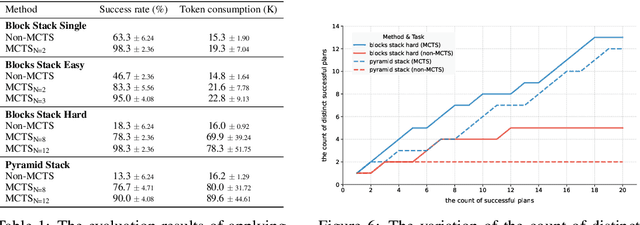
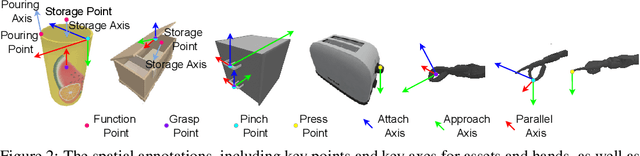
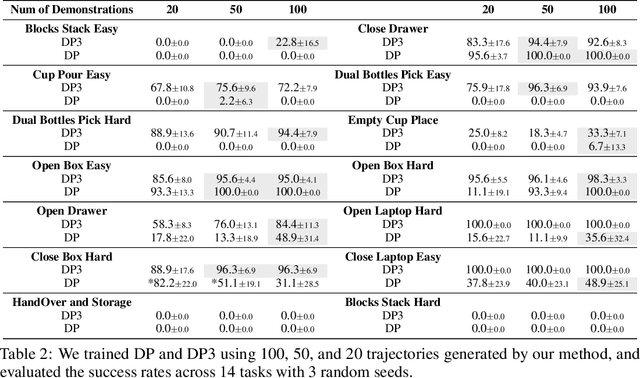
Abstract:For robotic manipulation, existing robotics datasets and simulation benchmarks predominantly cater to robot-arm platforms. However, for humanoid robots equipped with dual arms and dexterous hands, simulation tasks and high-quality demonstrations are notably lacking. Bimanual dexterous manipulation is inherently more complex, as it requires coordinated arm movements and hand operations, making autonomous data collection challenging. This paper presents HumanoidGen, an automated task creation and demonstration collection framework that leverages atomic dexterous operations and LLM reasoning to generate relational constraints. Specifically, we provide spatial annotations for both assets and dexterous hands based on the atomic operations, and perform an LLM planner to generate a chain of actionable spatial constraints for arm movements based on object affordances and scenes. To further improve planning ability, we employ a variant of Monte Carlo tree search to enhance LLM reasoning for long-horizon tasks and insufficient annotation. In experiments, we create a novel benchmark with augmented scenarios to evaluate the quality of the collected data. The results show that the performance of the 2D and 3D diffusion policies can scale with the generated dataset. Project page is https://openhumanoidgen.github.io.
Large Language Models on Fine-grained Emotion Detection Dataset with Data Augmentation and Transfer Learning
Mar 10, 2024Abstract:This paper delves into enhancing the classification performance on the GoEmotions dataset, a large, manually annotated dataset for emotion detection in text. The primary goal of this paper is to address the challenges of detecting subtle emotions in text, a complex issue in Natural Language Processing (NLP) with significant practical applications. The findings offer valuable insights into addressing the challenges of emotion detection in text and suggest directions for future research, including the potential for a survey paper that synthesizes methods and performances across various datasets in this domain.
Large Language Models for Forecasting and Anomaly Detection: A Systematic Literature Review
Feb 15, 2024



Abstract:This systematic literature review comprehensively examines the application of Large Language Models (LLMs) in forecasting and anomaly detection, highlighting the current state of research, inherent challenges, and prospective future directions. LLMs have demonstrated significant potential in parsing and analyzing extensive datasets to identify patterns, predict future events, and detect anomalous behavior across various domains. However, this review identifies several critical challenges that impede their broader adoption and effectiveness, including the reliance on vast historical datasets, issues with generalizability across different contexts, the phenomenon of model hallucinations, limitations within the models' knowledge boundaries, and the substantial computational resources required. Through detailed analysis, this review discusses potential solutions and strategies to overcome these obstacles, such as integrating multimodal data, advancements in learning methodologies, and emphasizing model explainability and computational efficiency. Moreover, this review outlines critical trends that are likely to shape the evolution of LLMs in these fields, including the push toward real-time processing, the importance of sustainable modeling practices, and the value of interdisciplinary collaboration. Conclusively, this review underscores the transformative impact LLMs could have on forecasting and anomaly detection while emphasizing the need for continuous innovation, ethical considerations, and practical solutions to realize their full potential.
When Large Language Models Meet Vector Databases: A Survey
Feb 06, 2024Abstract:This survey explores the synergistic potential of Large Language Models (LLMs) and Vector Databases (VecDBs), a burgeoning but rapidly evolving research area. With the proliferation of LLMs comes a host of challenges, including hallucinations, outdated knowledge, prohibitive commercial application costs, and memory issues. VecDBs emerge as a compelling solution to these issues by offering an efficient means to store, retrieve, and manage the high-dimensional vector representations intrinsic to LLM operations. Through this nuanced review, we delineate the foundational principles of LLMs and VecDBs and critically analyze their integration's impact on enhancing LLM functionalities. This discourse extends into a discussion on the speculative future developments in this domain, aiming to catalyze further research into optimizing the confluence of LLMs and VecDBs for advanced data handling and knowledge extraction capabilities.
Variational Autoencoder for Anti-Cancer Drug Response Prediction
Sep 29, 2020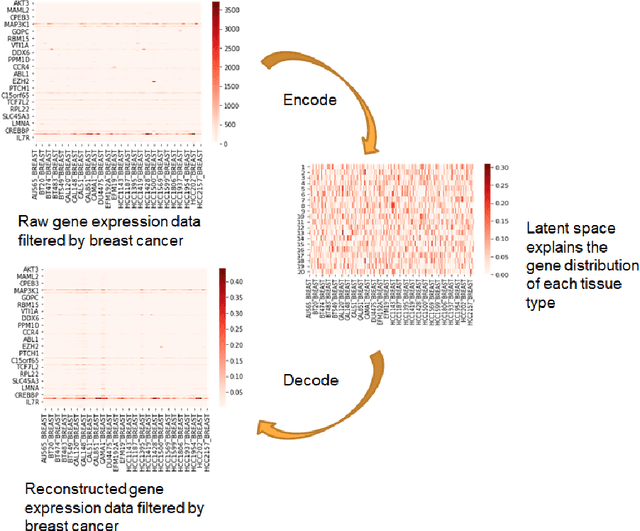

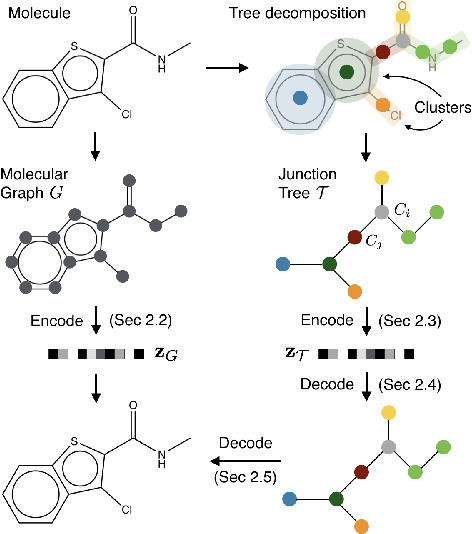
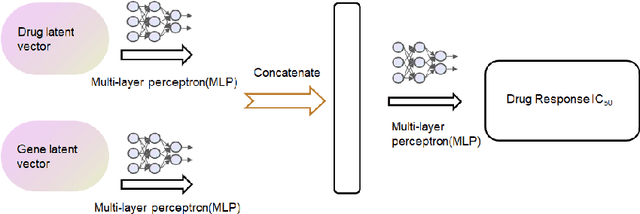
Abstract:Cancer has long been a main cause of human death, and the discovery of new drugs and the customization of cancer therapy have puzzled people for a long time. In order to facilitate the discovery of new anti-cancer drugs and the customization of treatment strategy, we seek to predict the response of different anti-cancer drugs with variational autoencoders (VAE) and multi-layer perceptron (MLP).Our model takes as input gene expression data of cancer cell lines and anti-cancer drug molecular data, and encode these data with {\sc {GeneVae}} model, which is an ordinary VAE, and rectified junction tree variational autoencoder ({\sc JtVae}) (\cite{jin2018junction}) model, respectively. Encoded features are processes by a Multi-layer Perceptron (MLP) model to produce a final prediction. We reach an average coefficient of determination ($R^{2} = 0.83$) in predicting drug response on breast cancer cell lines and an average $R^{2} > 0.84$ on pan-cancer cell lines. Additionally, we show that our model can generate unseen effective drug compounds for specific cancer cell lines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge