Yuxuan Tian
Scaling Up AI-Generated Image Detection via Generator-Aware Prototypes
Dec 15, 2025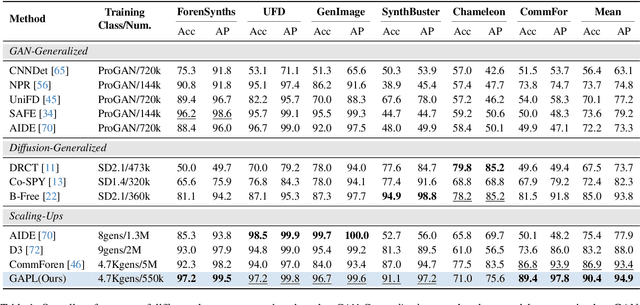
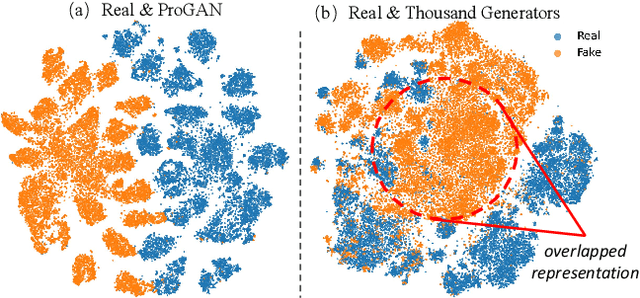
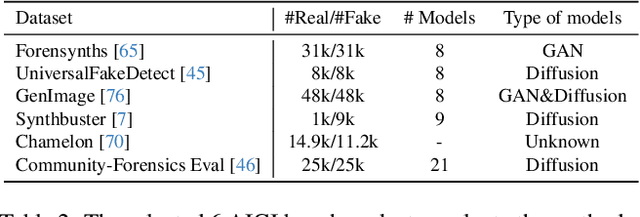
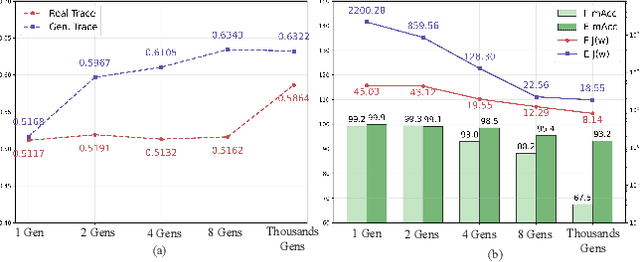
Abstract:The pursuit of a universal AI-generated image (AIGI) detector often relies on aggregating data from numerous generators to improve generalization. However, this paper identifies a paradoxical phenomenon we term the Benefit then Conflict dilemma, where detector performance stagnates and eventually degrades as source diversity expands. Our systematic analysis, diagnoses this failure by identifying two core issues: severe data-level heterogeneity, which causes the feature distributions of real and synthetic images to increasingly overlap, and a critical model-level bottleneck from fixed, pretrained encoders that cannot adapt to the rising complexity. To address these challenges, we propose Generator-Aware Prototype Learning (GAPL), a framework that constrain representation with a structured learning paradigm. GAPL learns a compact set of canonical forgery prototypes to create a unified, low-variance feature space, effectively countering data heterogeneity.To resolve the model bottleneck, it employs a two-stage training scheme with Low-Rank Adaptation, enhancing its discriminative power while preserving valuable pretrained knowledge. This approach establishes a more robust and generalizable decision boundary. Through extensive experiments, we demonstrate that GAPL achieves state-of-the-art performance, showing superior detection accuracy across a wide variety of GAN and diffusion-based generators. Code is available at https://github.com/UltraCapture/GAPL
DP-FedPGN: Finding Global Flat Minima for Differentially Private Federated Learning via Penalizing Gradient Norm
Oct 31, 2025Abstract:To prevent inference attacks in Federated Learning (FL) and reduce the leakage of sensitive information, Client-level Differentially Private Federated Learning (CL-DPFL) is widely used. However, current CL-DPFL methods usually result in sharper loss landscapes, which leads to a decrease in model generalization after differential privacy protection. By using Sharpness Aware Minimization (SAM), the current popular federated learning methods are to find a local flat minimum value to alleviate this problem. However, the local flatness may not reflect the global flatness in CL-DPFL. Therefore, to address this issue and seek global flat minima of models, we propose a new CL-DPFL algorithm, DP-FedPGN, in which we introduce a global gradient norm penalty to the local loss to find the global flat minimum. Moreover, by using our global gradient norm penalty, we not only find a flatter global minimum but also reduce the locally updated norm, which means that we further reduce the error of gradient clipping. From a theoretical perspective, we analyze how DP-FedPGN mitigates the performance degradation caused by DP. Meanwhile, the proposed DP-FedPGN algorithm eliminates the impact of data heterogeneity and achieves fast convergence. We also use R\'enyi DP to provide strict privacy guarantees and provide sensitivity analysis for local updates. Finally, we conduct effectiveness tests on both ResNet and Transformer models, and achieve significant improvements in six visual and natural language processing tasks compared to existing state-of-the-art algorithms. The code is available at https://github.com/junkangLiu0/DP-FedPGN
KeepKV: Eliminating Output Perturbation in KV Cache Compression for Efficient LLMs Inference
Apr 14, 2025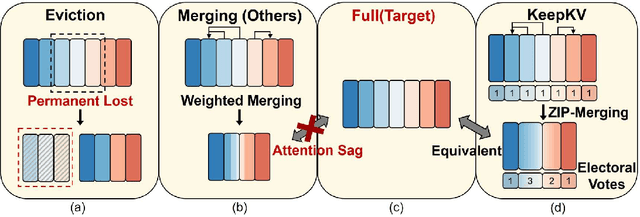
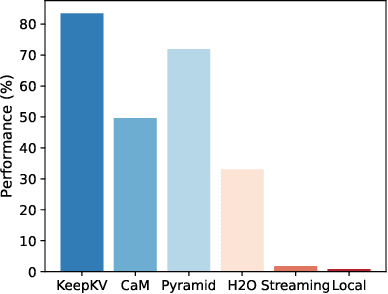
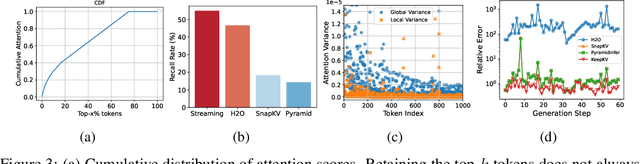
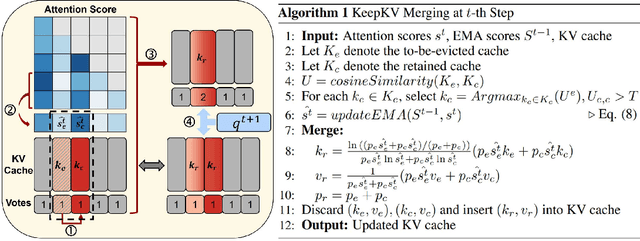
Abstract:Efficient inference of large language models (LLMs) is hindered by an ever-growing key-value (KV) cache, making KV cache compression a critical research direction. Traditional methods selectively evict less important KV cache entries based on attention scores or position heuristics, which leads to information loss and hallucinations. Recently, merging-based strategies have been explored to retain more information by merging KV pairs that would be discarded; however, these existing approaches inevitably introduce inconsistencies in attention distributions before and after merging, causing output perturbation and degraded generation quality. To overcome this challenge, we propose KeepKV, a novel adaptive KV cache merging method designed to eliminate output perturbation while preserving performance under strict memory constraints. KeepKV introduces the Electoral Votes mechanism that records merging history and adaptively adjusts attention scores. Moreover, it further leverages a novel Zero Inference-Perturbation Merging methods, keeping attention consistency and compensating for attention loss resulting from cache merging. KeepKV successfully retains essential context information within a significantly compressed cache. Extensive experiments on various benchmarks and LLM architectures demonstrate that KeepKV substantially reduces memory usage, enhances inference throughput by more than 2x and keeps superior generation quality even with 10% KV cache budgets.
FairKV: Balancing Per-Head KV Cache for Fast Multi-GPU Inference
Feb 19, 2025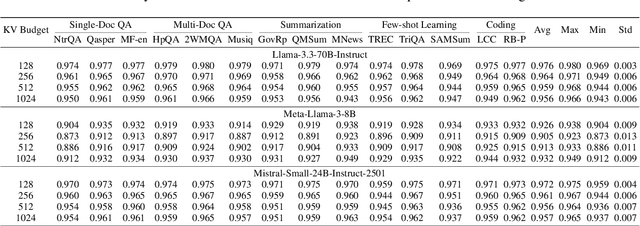
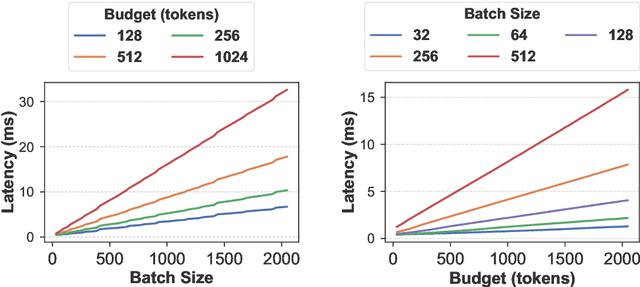
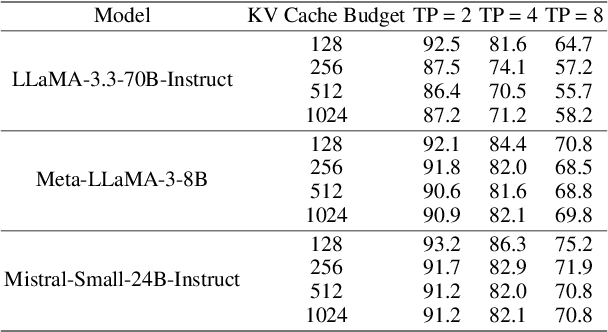
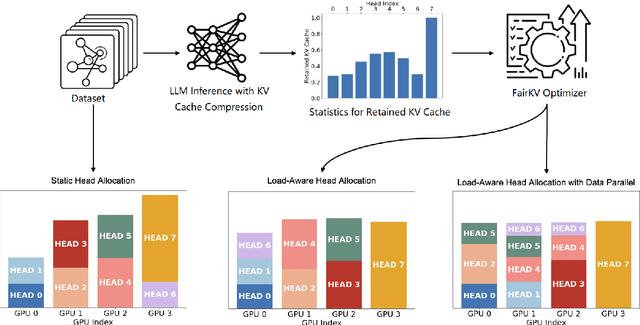
Abstract:KV cache techniques in Transformer models aim to reduce redundant computations at the expense of substantially increased memory usage, making KV cache compression an important and popular research topic. Recently, state-of-the-art KV cache compression methods implement imbalanced, per-head allocation algorithms that dynamically adjust the KV cache budget for each attention head, achieving excellent performance in single-GPU scenarios. However, we observe that such imbalanced compression leads to significant load imbalance when deploying multi-GPU inference, as some GPUs become overburdened while others remain underutilized. In this paper, we propose FairKV, a method designed to ensure fair memory usage among attention heads in systems employing imbalanced KV cache compression. The core technique of FairKV is Fair-Copying, which replicates a small subset of memory-intensive attention heads across GPUs using data parallelism to mitigate load imbalance. Our experiments on popular models, including LLaMA 70b and Mistral 24b model, demonstrate that FairKV increases throughput by 1.66x compared to standard tensor parallelism inference. Our code will be released as open source upon acceptance.
Prediction Is All MoE Needs: Expert Load Distribution Goes from Fluctuating to Stabilizing
Apr 25, 2024Abstract:MoE facilitates the development of large models by making the computational complexity of the model no longer scale linearly with increasing parameters. The learning sparse gating network selects a set of experts for each token to be processed; however, this may lead to differences in the number of tokens processed by each expert over several successive iterations, i.e., the expert load fluctuations, which reduces computational parallelization and resource utilization. To this end, we traced and analyzed loads of each expert in the training iterations for several large language models in this work, and defined the transient state with "obvious load fluctuation" and the stable state with "temporal locality". Moreover, given the characteristics of these two states and the computational overhead, we deployed three classical prediction algorithms that achieve accurate expert load prediction results. For the GPT3 350M model, the average error rates for predicting the expert load proportion over the next 1,000 and 2,000 steps are approximately 1.3% and 1.8%, respectively. This work can provide valuable guidance for expert placement or resource allocation for MoE model training. Based on this work, we will propose an expert placement scheme for transient and stable states in our coming work.
ProGroTrack: Deep Learning-Assisted Tracking of Intracellular Protein Growth Dynamics
May 26, 2023Abstract:Accurate tracking of cellular and subcellular structures, along with their dynamics, plays a pivotal role in understanding the underlying mechanisms of biological systems. This paper presents a novel approach, ProGroTrack, that combines the You Only Look Once (YOLO) and ByteTrack algorithms within the detection-based tracking (DBT) framework to track intracellular protein nanostructures. Focusing on iPAK4 protein fibers as a representative case study, we conducted a comprehensive evaluation of YOLOv5 and YOLOv8 models, revealing the superior performance of YOLOv5 on our dataset. Notably, YOLOv5x achieved an impressive mAP50 of 0.839 and F-score of 0.819. To further optimize detection capabilities, we incorporated semi-supervised learning for model improvement, resulting in enhanced performances in all metrics. Subsequently, we successfully applied our approach to track the growth behavior of iPAK4 protein fibers, revealing their two distinct growth phases consistent with a previously reported kinetic model. This research showcases the promising potential of our approach, extending beyond iPAK4 fibers. It also offers a significant advancement in precise tracking of dynamic processes in live cells, and fostering new avenues for biomedical research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge