Yulia Gel
When Witnesses Defend: A Witness Graph Topological Layer for Adversarial Graph Learning
Sep 24, 2024
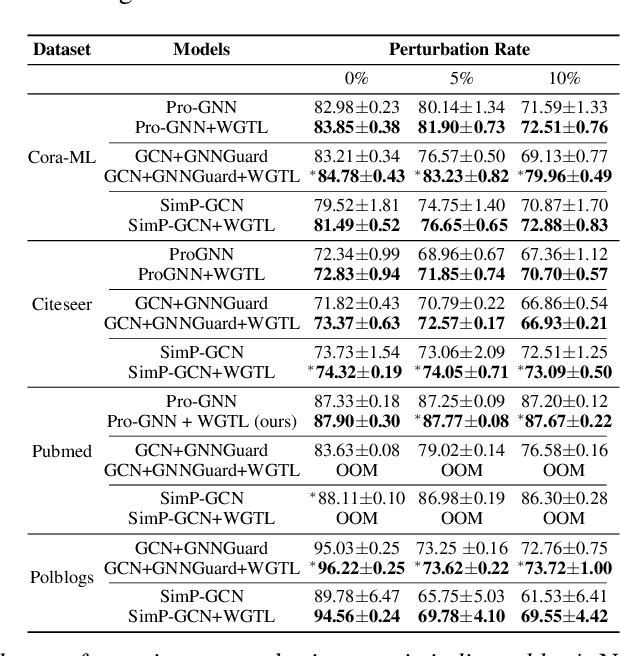
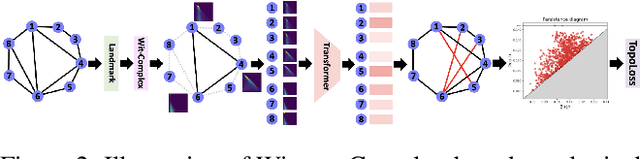
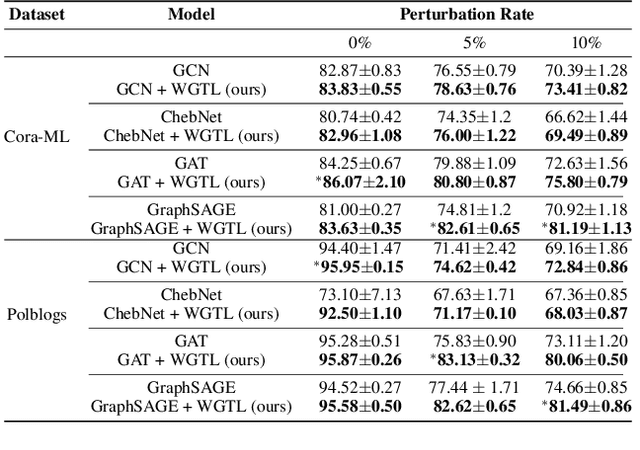
Abstract:Capitalizing on the intuitive premise that shape characteristics are more robust to perturbations, we bridge adversarial graph learning with the emerging tools from computational topology, namely, persistent homology representations of graphs. We introduce the concept of witness complex to adversarial analysis on graphs, which allows us to focus only on the salient shape characteristics of graphs, yielded by the subset of the most essential nodes (i.e., landmarks), with minimal loss of topological information on the whole graph. The remaining nodes are then used as witnesses, governing which higher-order graph substructures are incorporated into the learning process. Armed with the witness mechanism, we design Witness Graph Topological Layer (WGTL), which systematically integrates both local and global topological graph feature representations, the impact of which is, in turn, automatically controlled by the robust regularized topological loss. Given the attacker's budget, we derive the important stability guarantees of both local and global topology encodings and the associated robust topological loss. We illustrate the versatility and efficiency of WGTL by its integration with five GNNs and three existing non-topological defense mechanisms. Our extensive experiments across six datasets demonstrate that WGTL boosts the robustness of GNNs across a range of perturbations and against a range of adversarial attacks, leading to relative gains of up to 18%.
Efficient Planning of Multi-Robot Collective Transport using Graph Reinforcement Learning with Higher Order Topological Abstraction
Mar 15, 2023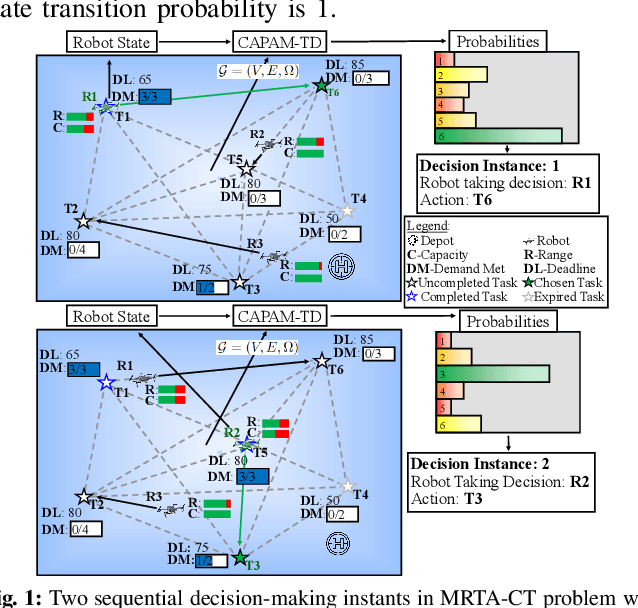
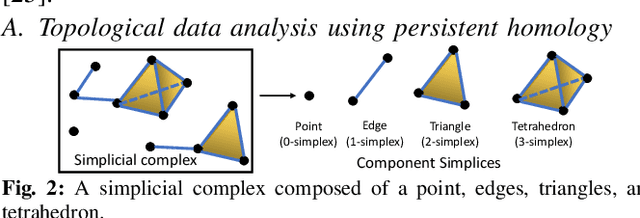

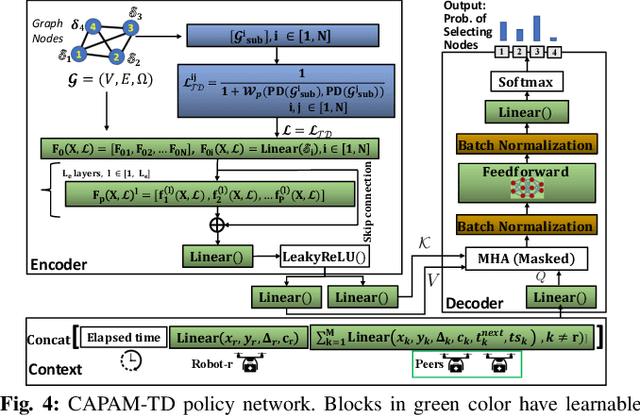
Abstract:Efficient multi-robot task allocation (MRTA) is fundamental to various time-sensitive applications such as disaster response, warehouse operations, and construction. This paper tackles a particular class of these problems that we call MRTA-collective transport or MRTA-CT -- here tasks present varying workloads and deadlines, and robots are subject to flight range, communication range, and payload constraints. For large instances of these problems involving 100s-1000's of tasks and 10s-100s of robots, traditional non-learning solvers are often time-inefficient, and emerging learning-based policies do not scale well to larger-sized problems without costly retraining. To address this gap, we use a recently proposed encoder-decoder graph neural network involving Capsule networks and multi-head attention mechanism, and innovatively add topological descriptors (TD) as new features to improve transferability to unseen problems of similar and larger size. Persistent homology is used to derive the TD, and proximal policy optimization is used to train our TD-augmented graph neural network. The resulting policy model compares favorably to state-of-the-art non-learning baselines while being much faster. The benefit of using TD is readily evident when scaling to test problems of size larger than those used in training.
ToDD: Topological Compound Fingerprinting in Computer-Aided Drug Discovery
Nov 07, 2022Abstract:In computer-aided drug discovery (CADD), virtual screening (VS) is used for identifying the drug candidates that are most likely to bind to a molecular target in a large library of compounds. Most VS methods to date have focused on using canonical compound representations (e.g., SMILES strings, Morgan fingerprints) or generating alternative fingerprints of the compounds by training progressively more complex variational autoencoders (VAEs) and graph neural networks (GNNs). Although VAEs and GNNs led to significant improvements in VS performance, these methods suffer from reduced performance when scaling to large virtual compound datasets. The performance of these methods has shown only incremental improvements in the past few years. To address this problem, we developed a novel method using multiparameter persistence (MP) homology that produces topological fingerprints of the compounds as multidimensional vectors. Our primary contribution is framing the VS process as a new topology-based graph ranking problem by partitioning a compound into chemical substructures informed by the periodic properties of its atoms and extracting their persistent homology features at multiple resolution levels. We show that the margin loss fine-tuning of pretrained Triplet networks attains highly competitive results in differentiating between compounds in the embedding space and ranking their likelihood of becoming effective drug candidates. We further establish theoretical guarantees for the stability properties of our proposed MP signatures, and demonstrate that our models, enhanced by the MP signatures, outperform state-of-the-art methods on benchmark datasets by a wide and highly statistically significant margin (e.g., 93% gain for Cleves-Jain and 54% gain for DUD-E Diverse dataset).
Unsupervised Space-Time Clustering using Persistent Homology
Oct 25, 2019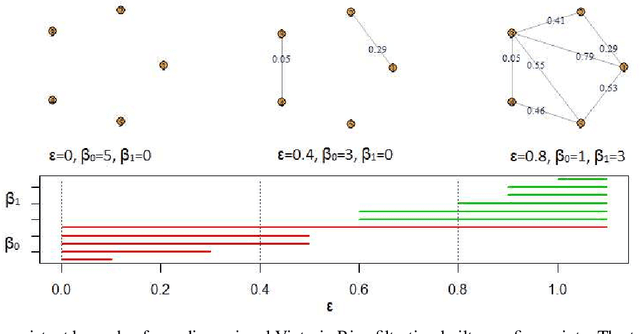
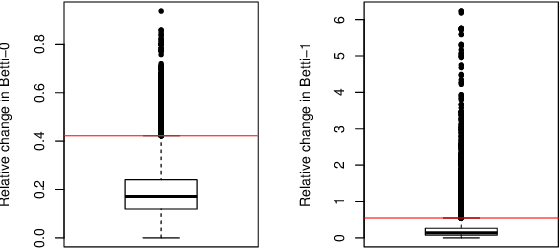
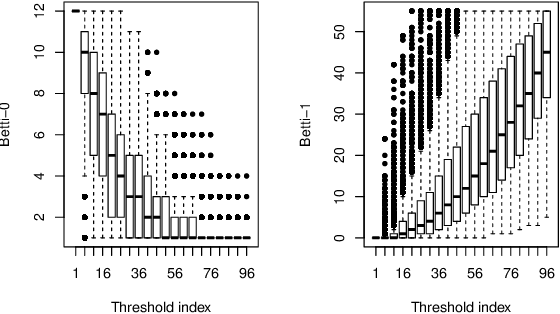
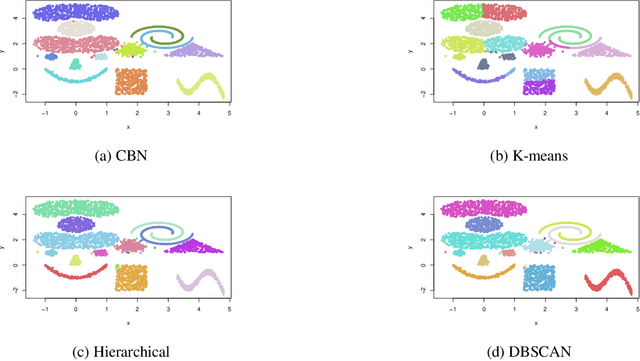
Abstract:This paper presents a new clustering algorithm for space-time data based on the concepts of topological data analysis and in particular, persistent homology. Employing persistent homology - a flexible mathematical tool from algebraic topology used to extract topological information from data - in unsupervised learning is an uncommon and a novel approach. A notable aspect of this methodology consists in analyzing data at multiple resolutions which allows to distinguish true features from noise based on the extent of their persistence. We evaluate the performance of our algorithm on synthetic data and compare it to other well-known clustering algorithms such as K-means, hierarchical clustering and DBSCAN. We illustrate its application in the context of a case study of water quality in the Chesapeake Bay.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge