Yuhua Zhou
GeoSketch: A Neural-Symbolic Approach to Geometric Multimodal Reasoning with Auxiliary Line Construction and Affine Transformation
Sep 26, 2025Abstract:Geometric Problem Solving (GPS) poses a unique challenge for Multimodal Large Language Models (MLLMs), requiring not only the joint interpretation of text and diagrams but also iterative visuospatial reasoning. While existing approaches process diagrams as static images, they lack the capacity for dynamic manipulation - a core aspect of human geometric reasoning involving auxiliary line construction and affine transformations. We present GeoSketch, a neural-symbolic framework that recasts geometric reasoning as an interactive perception-reasoning-action loop. GeoSketch integrates: (1) a Perception module that abstracts diagrams into structured logic forms, (2) a Symbolic Reasoning module that applies geometric theorems to decide the next deductive step, and (3) a Sketch Action module that executes operations such as drawing auxiliary lines or applying transformations, thereby updating the diagram in a closed loop. To train this agent, we develop a two-stage pipeline: supervised fine-tuning on 2,000 symbolic-curated trajectories followed by reinforcement learning with dense, symbolic rewards to enhance robustness and strategic exploration. To evaluate this paradigm, we introduce the GeoSketch Benchmark, a high-quality set of 390 geometry problems requiring auxiliary construction or affine transformations. Experiments on strong MLLM baselines demonstrate that GeoSketch significantly improves stepwise reasoning accuracy and problem-solving success over static perception methods. By unifying hierarchical decision-making, executable visual actions, and symbolic verification, GeoSketch advances multimodal reasoning from static interpretation to dynamic, verifiable interaction, establishing a new foundation for solving complex visuospatial problems.
Efficient Fine-Tuning of Quantized Models via Adaptive Rank and Bitwidth
May 02, 2025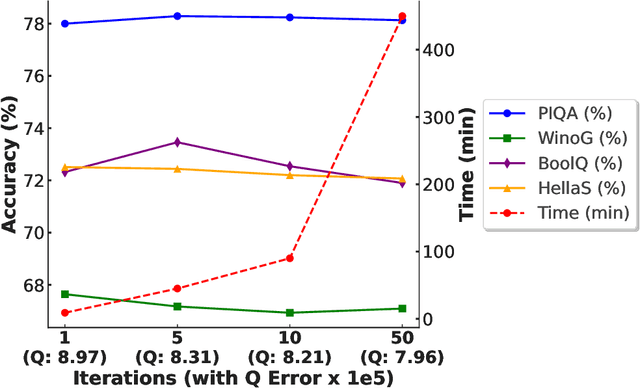
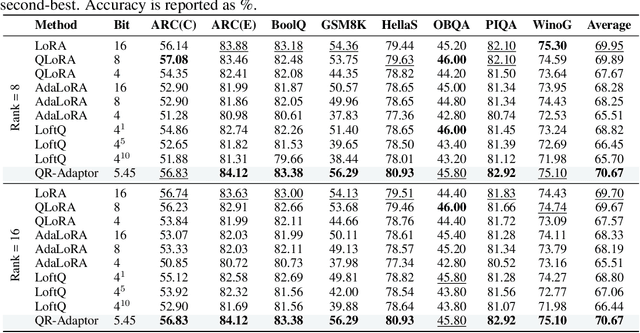
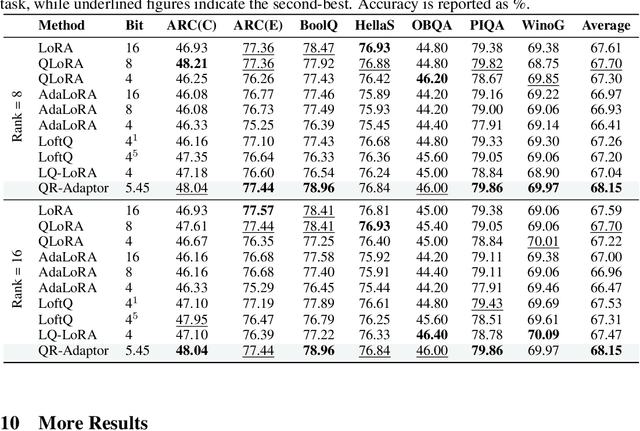
Abstract:QLoRA effectively combines low-bit quantization and LoRA to achieve memory-friendly fine-tuning for large language models (LLM). Recently, methods based on SVD for continuous update iterations to initialize LoRA matrices to accommodate quantization errors have generally failed to consistently improve performance. Dynamic mixed precision is a natural idea for continuously improving the fine-tuning performance of quantized models, but previous methods often optimize low-rank subspaces or quantization components separately, without considering their synergy. To address this, we propose \textbf{QR-Adaptor}, a unified, gradient-free strategy that uses partial calibration data to jointly search the quantization components and the rank of low-rank spaces for each layer, thereby continuously improving model performance. QR-Adaptor does not minimize quantization error but treats precision and rank allocation as a discrete optimization problem guided by actual downstream performance and memory usage. Compared to state-of-the-art (SOTA) quantized LoRA fine-tuning methods, our approach achieves a 4.89\% accuracy improvement on GSM8K, and in some cases even outperforms the 16-bit fine-tuned model while maintaining the memory footprint of the 4-bit setting.
UniMatch: Universal Matching from Atom to Task for Few-Shot Drug Discovery
Feb 18, 2025Abstract:Drug discovery is crucial for identifying candidate drugs for various diseases.However, its low success rate often results in a scarcity of annotations, posing a few-shot learning problem. Existing methods primarily focus on single-scale features, overlooking the hierarchical molecular structures that determine different molecular properties. To address these issues, we introduce Universal Matching Networks (UniMatch), a dual matching framework that integrates explicit hierarchical molecular matching with implicit task-level matching via meta-learning, bridging multi-level molecular representations and task-level generalization. Specifically, our approach explicitly captures structural features across multiple levels, such as atoms, substructures, and molecules, via hierarchical pooling and matching, facilitating precise molecular representation and comparison. Additionally, we employ a meta-learning strategy for implicit task-level matching, allowing the model to capture shared patterns across tasks and quickly adapt to new ones. This unified matching framework ensures effective molecular alignment while leveraging shared meta-knowledge for fast adaptation. Our experimental results demonstrate that UniMatch outperforms state-of-the-art methods on the MoleculeNet and FS-Mol benchmarks, achieving improvements of 2.87% in AUROC and 6.52% in delta AUPRC. UniMatch also shows excellent generalization ability on the Meta-MolNet benchmark.
QPruner: Probabilistic Decision Quantization for Structured Pruning in Large Language Models
Dec 16, 2024Abstract:The rise of large language models (LLMs) has significantly advanced various natural language processing (NLP) tasks. However, the resource demands of these models pose substantial challenges. Structured pruning is an effective approach to reducing model size, but it often results in significant accuracy degradation, necessitating parameter updates to adapt. Unfortunately, such fine-tuning requires substantial memory, which limits its applicability. To address these challenges, we introduce quantization into the structured pruning framework to reduce memory consumption during both fine-tuning and inference. However, the combined errors from pruning and quantization increase the difficulty of fine-tuning, requiring a more refined quantization scheme. To this end, we propose QPruner, a novel framework that employs structured pruning to reduce model size, followed by a layer-wise mixed-precision quantization scheme. Quantization precisions are assigned to each layer based on their importance to the target task, and Bayesian optimization is employed to refine precision allocation strategies, ensuring a balance between model accuracy and memory efficiency. Extensive experiments on benchmark datasets demonstrate that QPruner significantly outperforms existing methods in memory savings while maintaining or improving model performance.
AutoMixQ: Self-Adjusting Quantization for High Performance Memory-Efficient Fine-Tuning
Nov 21, 2024Abstract:Fine-tuning large language models (LLMs) under resource constraints is a significant challenge in deep learning. Low-Rank Adaptation (LoRA), pruning, and quantization are all effective methods for improving resource efficiency. However, combining them directly often results in suboptimal performance, especially with uniform quantization across all model layers. This is due to the complex, uneven interlayer relationships introduced by pruning, necessitating more refined quantization strategies. To address this, we propose AutoMixQ, an end-to-end optimization framework that selects optimal quantization configurations for each LLM layer. AutoMixQ leverages lightweight performance models to guide the selection process, significantly reducing time and computational resources compared to exhaustive search methods. By incorporating Pareto optimality, AutoMixQ balances memory usage and performance, approaching the upper bounds of model capability under strict resource constraints. Our experiments on widely used benchmarks show that AutoMixQ reduces memory consumption while achieving superior performance. For example, at a 30\% pruning rate in LLaMA-7B, AutoMixQ achieved 66.21\% on BoolQ compared to 62.45\% for LoRA and 58.96\% for LoftQ, while reducing memory consumption by 35.5\% compared to LoRA and 27.5\% compared to LoftQ.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge