Yugyung Lee
MATEX: Multi-scale Attention and Text-guided Explainability of Medical Vision-Language Models
Jan 16, 2026Abstract:We introduce MATEX (Multi-scale Attention and Text-guided Explainability), a novel framework that advances interpretability in medical vision-language models by incorporating anatomically informed spatial reasoning. MATEX synergistically combines multi-layer attention rollout, text-guided spatial priors, and layer consistency analysis to produce precise, stable, and clinically meaningful gradient attribution maps. By addressing key limitations of prior methods, such as spatial imprecision, lack of anatomical grounding, and limited attention granularity, MATEX enables more faithful and interpretable model explanations. Evaluated on the MS-CXR dataset, MATEX outperforms the state-of-the-art M2IB approach in both spatial precision and alignment with expert-annotated findings. These results highlight MATEX's potential to enhance trust and transparency in radiological AI applications.
Predicting When to Trust Vision-Language Models for Spatial Reasoning
Jan 14, 2026Abstract:Vision-Language Models (VLMs) demonstrate impressive capabilities across multimodal tasks, yet exhibit systematic spatial reasoning failures, achieving only 49% (CLIP) to 54% (BLIP-2) accuracy on basic directional relationships. For safe deployment in robotics and autonomous systems, we need to predict when to trust VLM spatial predictions rather than accepting all outputs. We propose a vision-based confidence estimation framework that validates VLM predictions through independent geometric verification using object detection. Unlike text-based approaches relying on self-assessment, our method fuses four signals via gradient boosting: geometric alignment between VLM claims and coordinates, spatial ambiguity from overlap, detection quality, and VLM internal uncertainty. We achieve 0.674 AUROC on BLIP-2 (34.0% improvement over text-based baselines) and 0.583 AUROC on CLIP (16.1% improvement), generalizing across generative and classification architectures. Our framework enables selective prediction: at 60% target accuracy, we achieve 61.9% coverage versus 27.6% baseline (2.2x improvement) on BLIP-2. Feature analysis reveals vision-based signals contribute 87.4% of model importance versus 12.7% from VLM confidence, validating that external geometric verification outperforms self-assessment. We demonstrate reliable scene graph construction where confidence-based pruning improves precision from 52.1% to 78.3% while retaining 68.2% of edges.
Clinical Knowledge Graph Construction and Evaluation with Multi-LLMs via Retrieval-Augmented Generation
Jan 05, 2026Abstract:Large language models (LLMs) offer new opportunities for constructing knowledge graphs (KGs) from unstructured clinical narratives. However, existing approaches often rely on structured inputs and lack robust validation of factual accuracy and semantic consistency, limitations that are especially problematic in oncology. We introduce an end-to-end framework for clinical KG construction and evaluation directly from free text using multi-agent prompting and a schema-constrained Retrieval-Augmented Generation (KG-RAG) strategy. Our pipeline integrates (1) prompt-driven entity, attribute, and relation extraction; (2) entropy-based uncertainty scoring; (3) ontology-aligned RDF/OWL schema generation; and (4) multi-LLM consensus validation for hallucination detection and semantic refinement. Beyond static graph construction, the framework supports continuous refinement and self-supervised evaluation, enabling iterative improvement of graph quality. Applied to two oncology cohorts (PDAC and BRCA), our method produces interpretable, SPARQL-compatible, and clinically grounded knowledge graphs without relying on gold-standard annotations. Experimental results demonstrate consistent gains in precision, relevance, and ontology compliance over baseline methods.
Multi-Modal Interpretability for Enhanced Localization in Vision-Language Models
Sep 17, 2025Abstract:Recent advances in vision-language models have significantly expanded the frontiers of automated image analysis. However, applying these models in safety-critical contexts remains challenging due to the complex relationships between objects, subtle visual cues, and the heightened demand for transparency and reliability. This paper presents the Multi-Modal Explainable Learning (MMEL) framework, designed to enhance the interpretability of vision-language models while maintaining high performance. Building upon prior work in gradient-based explanations for transformer architectures (Grad-eclip), MMEL introduces a novel Hierarchical Semantic Relationship Module that enhances model interpretability through multi-scale feature processing, adaptive attention weighting, and cross-modal alignment. Our approach processes features at multiple semantic levels to capture relationships between image regions at different granularities, applying learnable layer-specific weights to balance contributions across the model's depth. This results in more comprehensive visual explanations that highlight both primary objects and their contextual relationships with improved precision. Through extensive experiments on standard datasets, we demonstrate that by incorporating semantic relationship information into gradient-based attribution maps, MMEL produces more focused and contextually aware visualizations that better reflect how vision-language models process complex scenes. The MMEL framework generalizes across various domains, offering valuable insights into model decisions for applications requiring high interpretability and reliability.
* 8 pages, 6 figures, 3 tables
OCU-Net: A Novel U-Net Architecture for Enhanced Oral Cancer Segmentation
Oct 03, 2023



Abstract:Accurate detection of oral cancer is crucial for improving patient outcomes. However, the field faces two key challenges: the scarcity of deep learning-based image segmentation research specifically targeting oral cancer and the lack of annotated data. Our study proposes OCU-Net, a pioneering U-Net image segmentation architecture exclusively designed to detect oral cancer in hematoxylin and eosin (H&E) stained image datasets. OCU-Net incorporates advanced deep learning modules, such as the Channel and Spatial Attention Fusion (CSAF) module, a novel and innovative feature that emphasizes important channel and spatial areas in H&E images while exploring contextual information. In addition, OCU-Net integrates other innovative components such as Squeeze-and-Excite (SE) attention module, Atrous Spatial Pyramid Pooling (ASPP) module, residual blocks, and multi-scale fusion. The incorporation of these modules showed superior performance for oral cancer segmentation for two datasets used in this research. Furthermore, we effectively utilized the efficient ImageNet pre-trained MobileNet-V2 model as a backbone of our OCU-Net to create OCU-Netm, an enhanced version achieving state-of-the-art results. Comprehensive evaluation demonstrates that OCU-Net and OCU-Netm outperformed existing segmentation methods, highlighting their precision in identifying cancer cells in H&E images from OCDC and ORCA datasets.
EARLIN: Early Out-of-Distribution Detection for Resource-efficient Collaborative Inference
Jun 29, 2021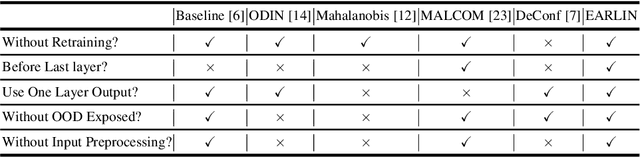

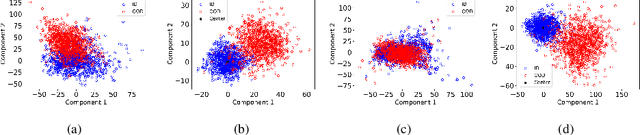
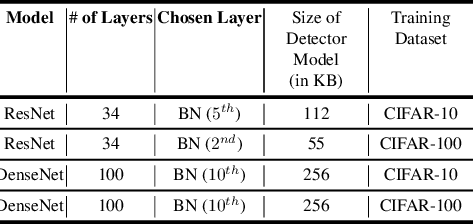
Abstract:Collaborative inference enables resource-constrained edge devices to make inferences by uploading inputs (e.g., images) to a server (i.e., cloud) where the heavy deep learning models run. While this setup works cost-effectively for successful inferences, it severely underperforms when the model faces input samples on which the model was not trained (known as Out-of-Distribution (OOD) samples). If the edge devices could, at least, detect that an input sample is an OOD, that could potentially save communication and computation resources by not uploading those inputs to the server for inference workload. In this paper, we propose a novel lightweight OOD detection approach that mines important features from the shallow layers of a pretrained CNN model and detects an input sample as ID (In-Distribution) or OOD based on a distance function defined on the reduced feature space. Our technique (a) works on pretrained models without any retraining of those models, and (b) does not expose itself to any OOD dataset (all detection parameters are obtained from the ID training dataset). To this end, we develop EARLIN (EARLy OOD detection for Collaborative INference) that takes a pretrained model and partitions the model at the OOD detection layer and deploys the considerably small OOD part on an edge device and the rest on the cloud. By experimenting using real datasets and a prototype implementation, we show that our technique achieves better results than other approaches in terms of overall accuracy and cost when tested against popular OOD datasets on top of popular deep learning models pretrained on benchmark datasets.
Link Prediction for Temporally Consistent Networks
Jun 06, 2020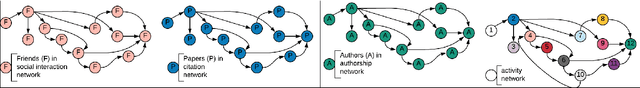
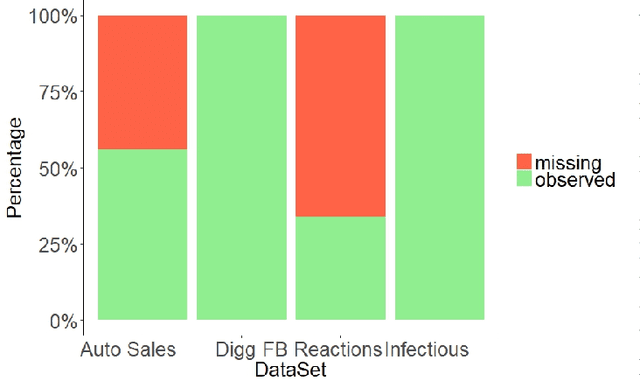


Abstract:Dynamic networks have intrinsic structural, computational, and multidisciplinary advantages. Link prediction estimates the next relationship in dynamic networks. However, in the current link prediction approaches, only bipartite or non-bipartite but homogeneous networks are considered. The use of adjacency matrix to represent dynamically evolving networks limits the ability to analytically learn from heterogeneous, sparse, or forming networks. In the case of a heterogeneous network, modeling all network states using a binary-valued matrix can be difficult. On the other hand, sparse or currently forming networks have many missing edges, which are represented as zeros, thus introducing class imbalance or noise. We propose a time-parameterized matrix (TP-matrix) and empirically demonstrate its effectiveness in non-bipartite, heterogeneous networks. In addition, we propose a predictive influence index as a measure of a node's boosting or diminishing predictive influence using backward and forward-looking maximization over the temporal space of the n-degree neighborhood. We further propose a new method of canonically representing heterogeneous time-evolving activities as a temporally parameterized network model (TPNM). The new method robustly enables activities to be represented as a form of a network, thus potentially inspiring new link prediction applications, including intelligent business process management systems and context-aware workflow engines. We evaluated our model on four datasets of different network systems. We present results that show the proposed model is more effective in capturing and retaining temporal relationships in dynamically evolving networks. We also show that our model performed better than state-of-the-art link prediction benchmark results for networks that are sensitive to temporal evolution.
SCAT: Second Chance Autoencoder for Textual Data
May 11, 2020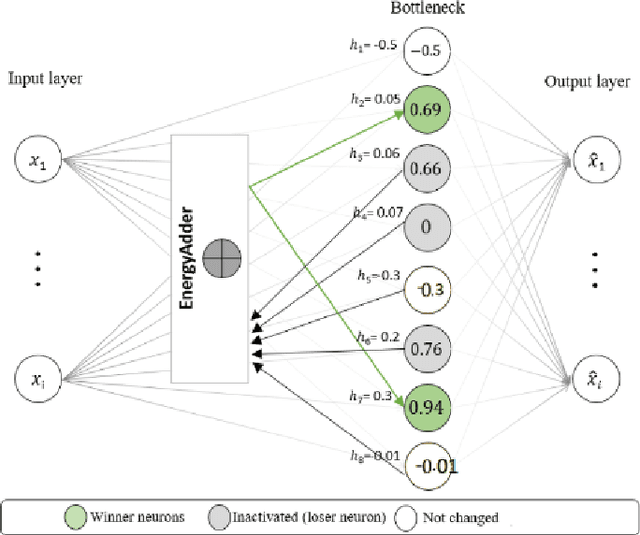
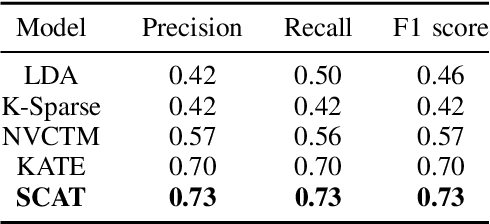
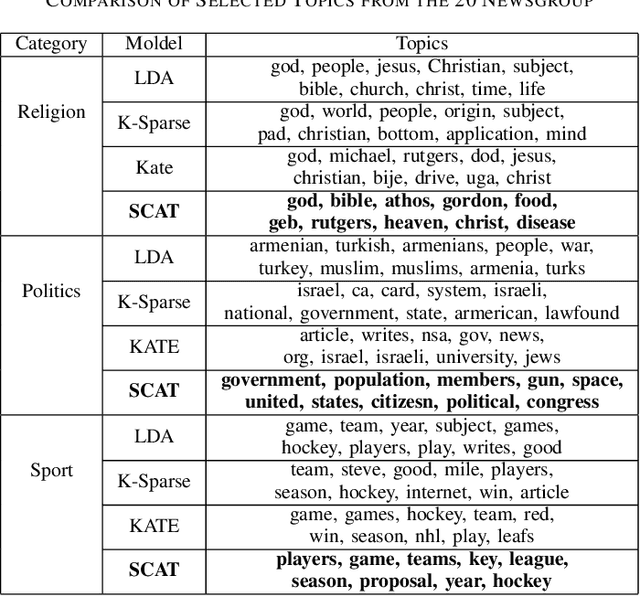
Abstract:We present a k-competitive learning approach for textual autoencoders named Second Chance Autoencoder (SCAT). SCAT selects the $k$ largest and smallest positive activations as the winner neurons, which gain the activation values of the loser neurons during the learning process, and thus focus on retrieving well-representative features for topics. Our experiments show that SCAT achieves outstanding performance in classification, topic modeling, and document visualization compared to LDA, K-Sparse, NVCTM, and KATE.
CRL: Class Representative Learning for Image Classification
Feb 16, 2020



Abstract:Building robust and real-time classifiers with diverse datasets are one of the most significant challenges to deep learning researchers. It is because there is a considerable gap between a model built with training (seen) data and real (unseen) data in applications. Recent works including Zero-Shot Learning (ZSL), have attempted to deal with this problem of overcoming the apparent gap through transfer learning. In this paper, we propose a novel model, called Class Representative Learning Model (CRL), that can be especially effective in image classification influenced by ZSL. In the CRL model, first, the learning step is to build class representatives to represent classes in datasets by aggregating prominent features extracted from a Convolutional Neural Network (CNN). Second, the inferencing step in CRL is to match between the class representatives and new data. The proposed CRL model demonstrated superior performance compared to the current state-of-the-art research in ZSL and mobile deep learning. The proposed CRL model has been implemented and evaluated in a parallel environment, using Apache Spark, for both distributed learning and recognition. An extensive experimental study on the benchmark datasets, ImageNet-1K, CalTech-101, CalTech-256, CIFAR-100, shows that CRL can build a class distribution model with drastic improvement in learning and recognition performance without sacrificing accuracy compared to the state-of-the-art performances in image classification.
Automated Human Claustrum Segmentation using Deep Learning Technologies
Nov 18, 2019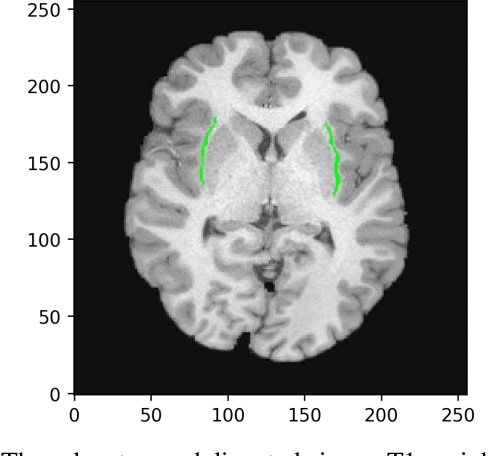
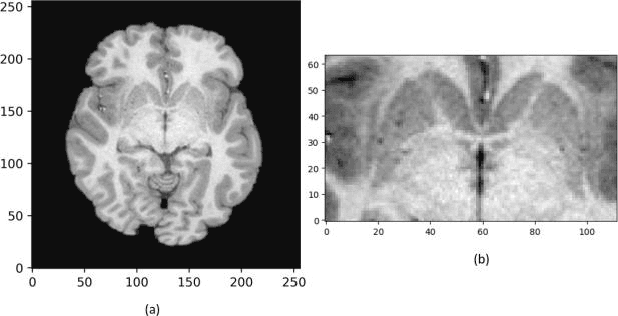
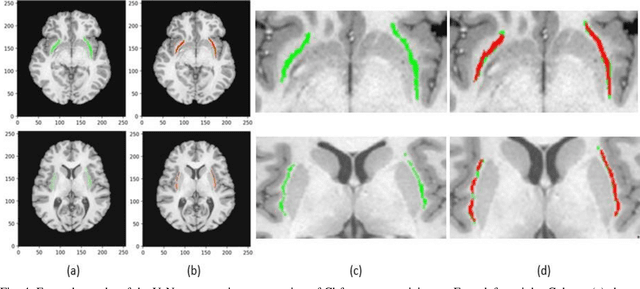
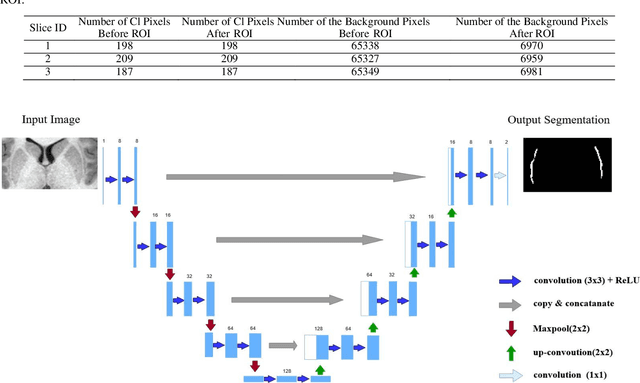
Abstract:In recent years, Deep Learning (DL) has shown promising results in conducting AI tasks such as computer vision and image segmentation. Specifically, Convolutional Neural Network (CNN) models in DL have been applied to prevention,detection, and diagnosis in predictive medicine. Image segmentation plays a significant role in disease detection and prevention.However, there are enormous challenges in performing DL-based automatic segmentation due to the nature of medical images such as heterogeneous modalities and formats, insufficient labeled training data, and the high-class imbalance in the labeled data. Furthermore, automating segmentation of medical images,like magnetic resonance images (MRI), becomes a challenging task. The need for automated segmentation or annotation is what motivates our work. In this paper, we propose a fully automated approach that aims to segment the human claustrum for analytical purposes. We applied a U-Net CNN model to segment the claustrum (Cl) from a MRI dataset. With this approach, we have achieved an average Dice per case score of 0.72 for Cl segmentation, with K=5 for cross-validation. The expert in the medical domain also evaluates these results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge