MedTQ: Dynamic Topic Discovery and Query Generation for Medical Ontologies
Paper and Code
Feb 12, 2018
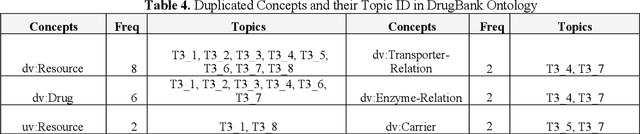
Biomedical ontology refers to a shared conceptualization for a biomedical domain of interest that has vastly improved data management and data sharing through the open data movement. The rapid growth and availability of biomedical data make it impractical and computationally expensive to perform manual analysis and query processing with the large scale ontologies. The lack of ability in analyzing ontologies from such a variety of sources, and supporting knowledge discovery for clinical practice and biomedical research should be overcome with new technologies. In this study, we developed a Medical Topic discovery and Query generation framework (MedTQ), which was composed by a series of approaches and algorithms. A predicate neighborhood pattern-based approach introduced has the ability to compute the similarity of predicates (relations) in ontologies. Given a predicate similarity metric, machine learning algorithms have been developed for automatic topic discovery and query generation. The topic discovery algorithm, called the hierarchical K-Means algorithm was designed by extending an existing supervised algorithm (K-means clustering) for the construction of a topic hierarchy. In the hierarchical K-Means algorithm, a level-by-level optimization strategy was selected for consistent with the strongly association between elements within a topic. Automatic query generation was facilitated for discovered topic that could be guided users for interactive query design and processing. Evaluation was conducted to generate topic hierarchy for DrugBank ontology as a case study. Results demonstrated that the MedTQ framework can enhance knowledge discovery by capturing underlying structures from domain specific data and ontologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge