Yiding Lu
LLaVA-ReID: Selective Multi-image Questioner for Interactive Person Re-Identification
Apr 15, 2025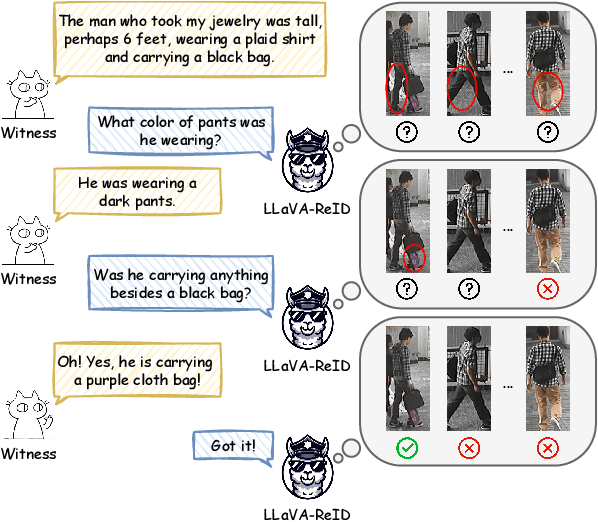
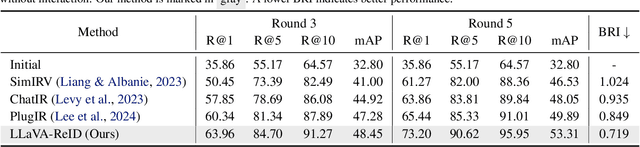
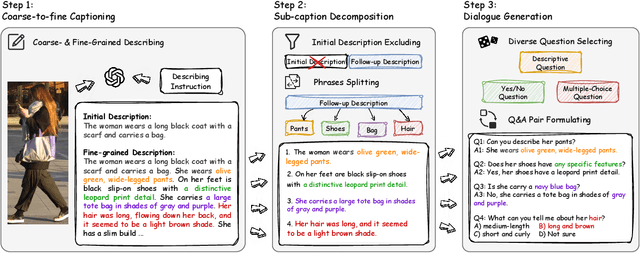
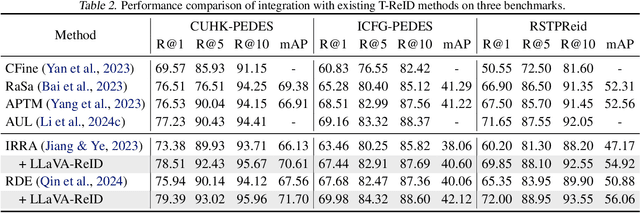
Abstract:Traditional text-based person ReID assumes that person descriptions from witnesses are complete and provided at once. However, in real-world scenarios, such descriptions are often partial or vague. To address this limitation, we introduce a new task called interactive person re-identification (Inter-ReID). Inter-ReID is a dialogue-based retrieval task that iteratively refines initial descriptions through ongoing interactions with the witnesses. To facilitate the study of this new task, we construct a dialogue dataset that incorporates multiple types of questions by decomposing fine-grained attributes of individuals. We further propose LLaVA-ReID, a question model that generates targeted questions based on visual and textual contexts to elicit additional details about the target person. Leveraging a looking-forward strategy, we prioritize the most informative questions as supervision during training. Experimental results on both Inter-ReID and text-based ReID benchmarks demonstrate that LLaVA-ReID significantly outperforms baselines.
A Survey on Deep Clustering: From the Prior Perspective
Jun 28, 2024Abstract:Facilitated by the powerful feature extraction ability of neural networks, deep clustering has achieved great success in analyzing high-dimensional and complex real-world data. The performance of deep clustering methods is affected by various factors such as network structures and learning objectives. However, as pointed out in this survey, the essence of deep clustering lies in the incorporation and utilization of prior knowledge, which is largely ignored by existing works. From pioneering deep clustering methods based on data structure assumptions to recent contrastive clustering methods based on data augmentation invariances, the development of deep clustering intrinsically corresponds to the evolution of prior knowledge. In this survey, we provide a comprehensive review of deep clustering methods by categorizing them into six types of prior knowledge. We find that in general the prior innovation follows two trends, namely, i) from mining to constructing, and ii) from internal to external. Besides, we provide a benchmark on five widely-used datasets and analyze the performance of methods with diverse priors. By providing a novel prior knowledge perspective, we hope this survey could provide some novel insights and inspire future research in the deep clustering community.
Decoupled Contrastive Multi-view Clustering with High-order Random Walks
Aug 22, 2023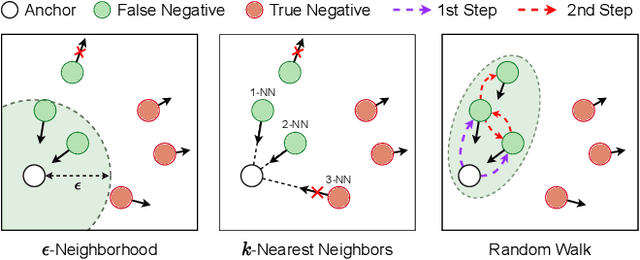
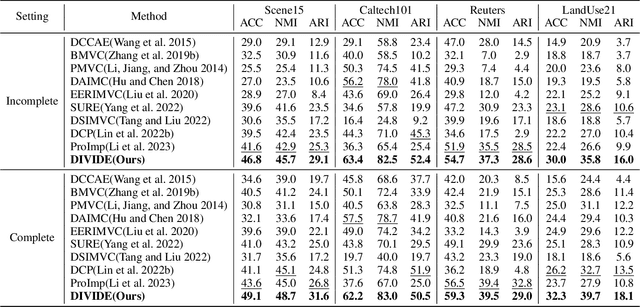
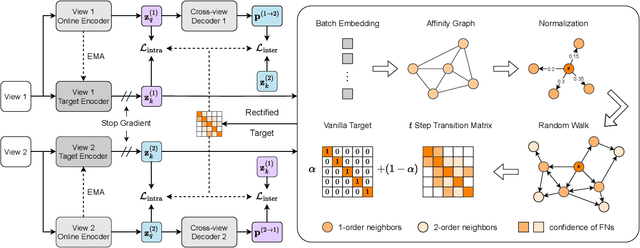
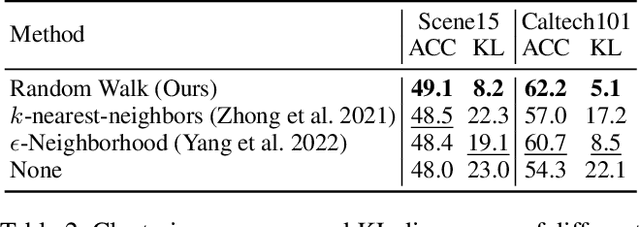
Abstract:In recent, some robust contrastive multi-view clustering (MvC) methods have been proposed, which construct data pairs from neighborhoods to alleviate the false negative issue, i.e., some intra-cluster samples are wrongly treated as negative pairs. Although promising performance has been achieved by these methods, the false negative issue is still far from addressed and the false positive issue emerges because all in- and out-of-neighborhood samples are simply treated as positive and negative, respectively. To address the issues, we propose a novel robust method, dubbed decoupled contrastive multi-view clustering with high-order random walks (DIVIDE). In brief, DIVIDE leverages random walks to progressively identify data pairs in a global instead of local manner. As a result, DIVIDE could identify in-neighborhood negatives and out-of-neighborhood positives. Moreover, DIVIDE embraces a novel MvC architecture to perform inter- and intra-view contrastive learning in different embedding spaces, thus boosting clustering performance and embracing the robustness against missing views. To verify the efficacy of DIVIDE, we carry out extensive experiments on four benchmark datasets comparing with nine state-of-the-art MvC methods in both complete and incomplete MvC settings.
Semantic Invariant Multi-view Clustering with Fully Incomplete Information
May 22, 2023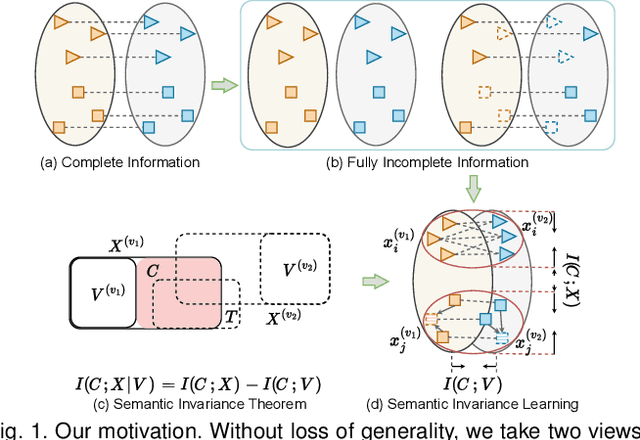
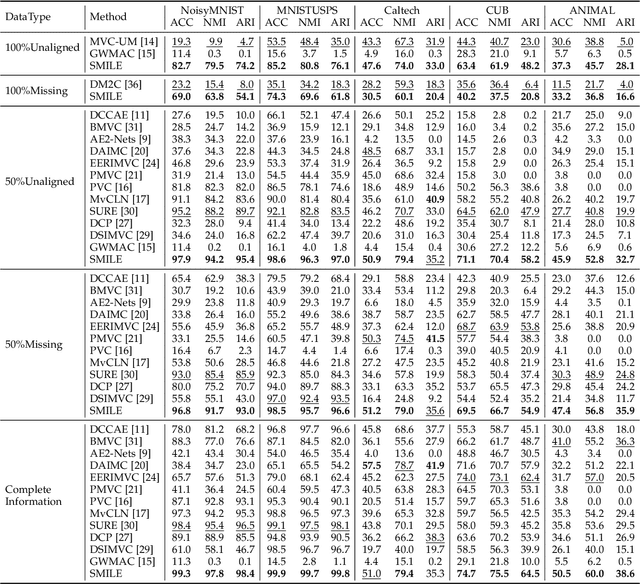
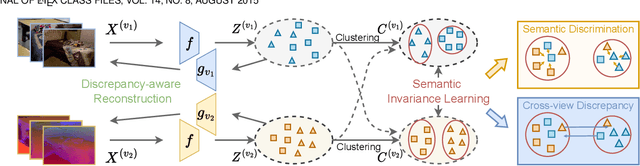
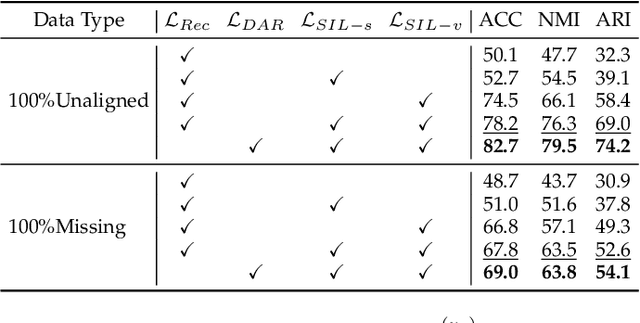
Abstract:Robust multi-view learning with incomplete information has received significant attention due to issues such as incomplete correspondences and incomplete instances that commonly affect real-world multi-view applications. Existing approaches heavily rely on paired samples to realign or impute defective ones, but such preconditions cannot always be satisfied in practice due to the complexity of data collection and transmission. To address this problem, we present a novel framework called SeMantic Invariance LEarning (SMILE) for multi-view clustering with incomplete information that does not require any paired samples. To be specific, we discover the existence of invariant semantic distribution across different views, which enables SMILE to alleviate the cross-view discrepancy to learn consensus semantics without requiring any paired samples. The resulting consensus semantics remains unaffected by cross-view distribution shifts, making them useful for realigning/imputing defective instances and forming clusters. We demonstrate the effectiveness of SMILE through extensive comparison experiments with 13 state-of-the-art baselines on five benchmarks. Our approach improves the clustering accuracy of NoisyMNIST from 19.3\%/23.2\% to 82.7\%/69.0\% when the correspondences/instances are fully incomplete. We will release the code after acceptance.
Erratum: Link prediction in drug-target interactions network using similarity indices
Nov 01, 2017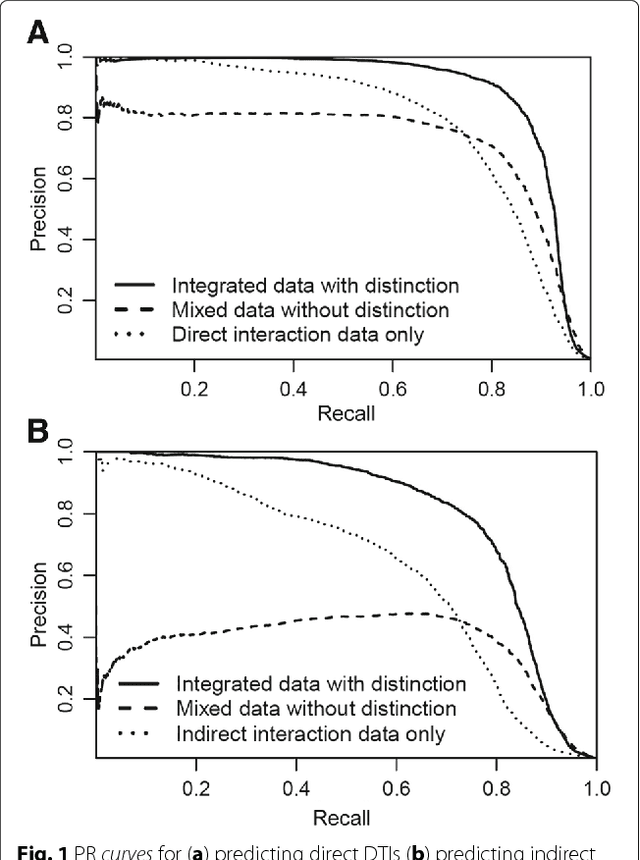

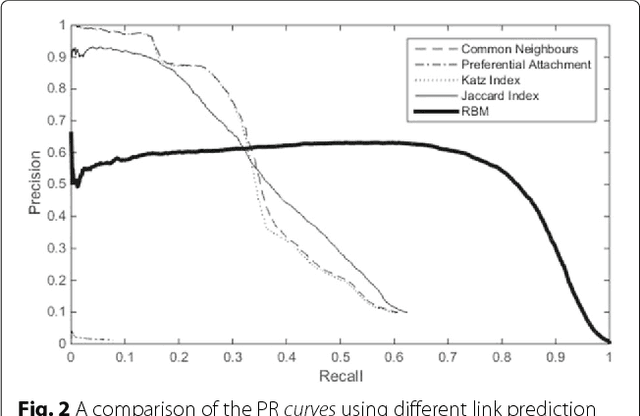
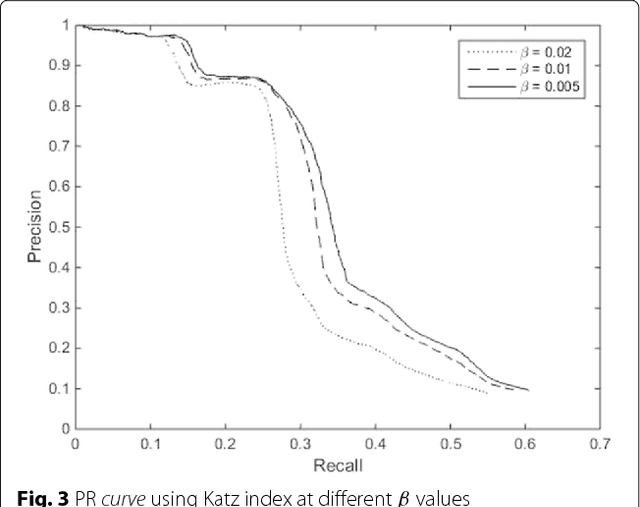
Abstract:Background: In silico drug-target interaction (DTI) prediction plays an integral role in drug repositioning: the discovery of new uses for existing drugs. One popular method of drug repositioning is network-based DTI prediction, which uses complex network theory to predict DTIs from a drug-target network. Currently, most network-based DTI prediction is based on machine learning methods such as Restricted Boltzmann Machines (RBM) or Support Vector Machines (SVM). These methods require additional information about the characteristics of drugs, targets and DTIs, such as chemical structure, genome sequence, binding types, causes of interactions, etc., and do not perform satisfactorily when such information is unavailable. We propose a new, alternative method for DTI prediction that makes use of only network topology information attempting to solve this problem. Results: We compare our method for DTI prediction against the well-known RBM approach. We show that when applied to the MATADOR database, our approach based on node neighborhoods yield higher precision for high-ranking predictions than RBM when no information regarding DTI types is available. Conclusion: This demonstrates that approaches purely based on network topology provide a more suitable approach to DTI prediction in the many real-life situations where little or no prior knowledge is available about the characteristics of drugs, targets, or their interactions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge