Erratum: Link prediction in drug-target interactions network using similarity indices
Paper and Code
Nov 01, 2017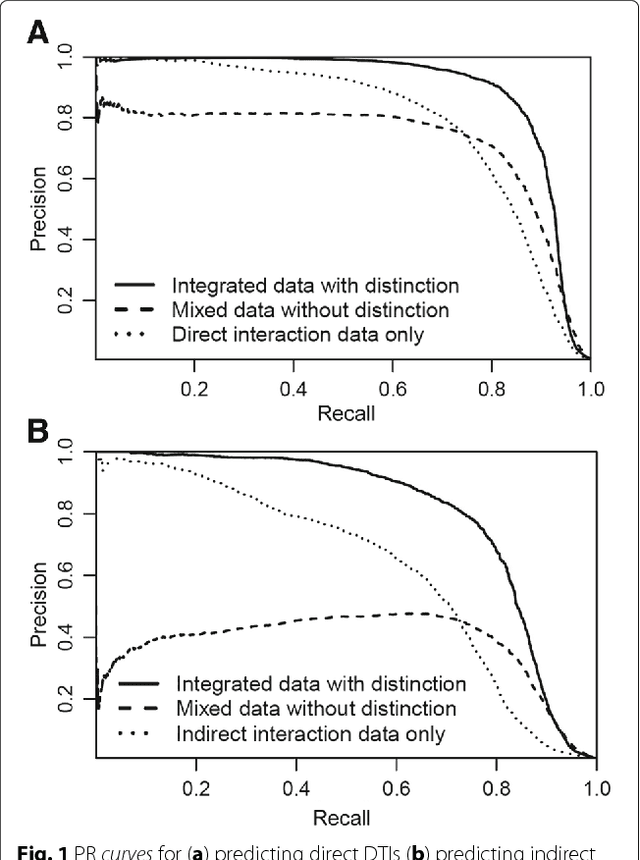

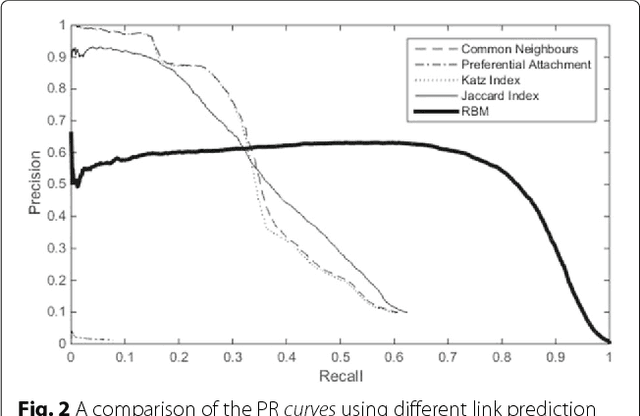
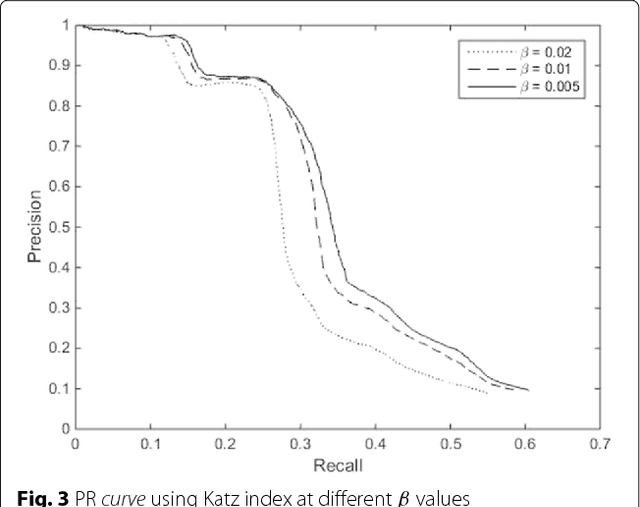
Background: In silico drug-target interaction (DTI) prediction plays an integral role in drug repositioning: the discovery of new uses for existing drugs. One popular method of drug repositioning is network-based DTI prediction, which uses complex network theory to predict DTIs from a drug-target network. Currently, most network-based DTI prediction is based on machine learning methods such as Restricted Boltzmann Machines (RBM) or Support Vector Machines (SVM). These methods require additional information about the characteristics of drugs, targets and DTIs, such as chemical structure, genome sequence, binding types, causes of interactions, etc., and do not perform satisfactorily when such information is unavailable. We propose a new, alternative method for DTI prediction that makes use of only network topology information attempting to solve this problem. Results: We compare our method for DTI prediction against the well-known RBM approach. We show that when applied to the MATADOR database, our approach based on node neighborhoods yield higher precision for high-ranking predictions than RBM when no information regarding DTI types is available. Conclusion: This demonstrates that approaches purely based on network topology provide a more suitable approach to DTI prediction in the many real-life situations where little or no prior knowledge is available about the characteristics of drugs, targets, or their interactions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge