Yi Lai
SCOPE-DTI: Semi-Inductive Dataset Construction and Framework Optimization for Practical Usability Enhancement in Deep Learning-Based Drug Target Interaction Prediction
Mar 12, 2025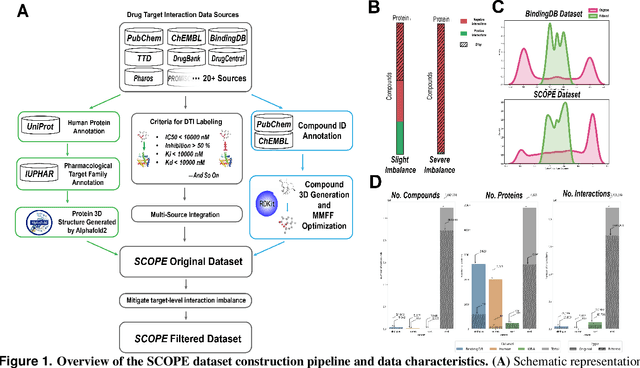
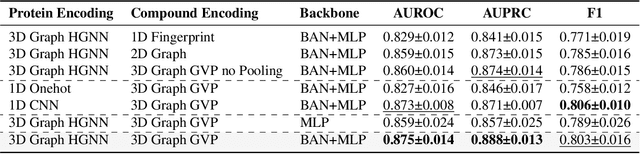
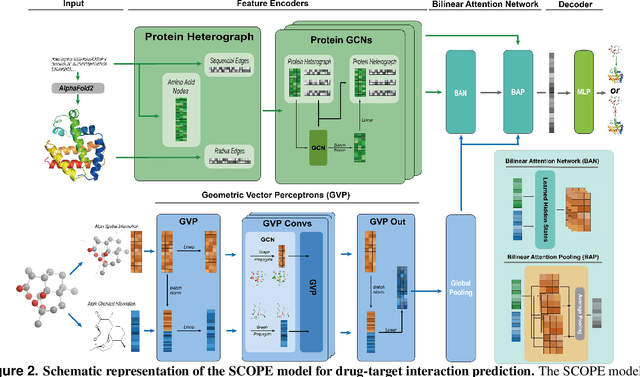
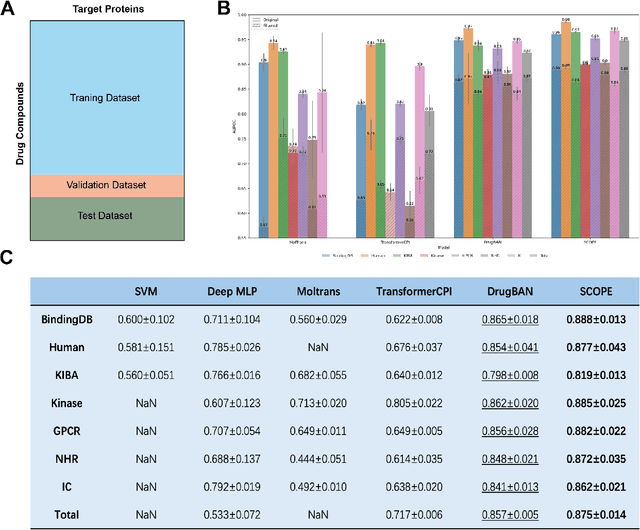
Abstract:Deep learning-based drug-target interaction (DTI) prediction methods have demonstrated strong performance; however, real-world applicability remains constrained by limited data diversity and modeling complexity. To address these challenges, we propose SCOPE-DTI, a unified framework combining a large-scale, balanced semi-inductive human DTI dataset with advanced deep learning modeling. Constructed from 13 public repositories, the SCOPE dataset expands data volume by up to 100-fold compared to common benchmarks such as the Human dataset. The SCOPE model integrates three-dimensional protein and compound representations, graph neural networks, and bilinear attention mechanisms to effectively capture cross domain interaction patterns, significantly outperforming state-of-the-art methods across various DTI prediction tasks. Additionally, SCOPE-DTI provides a user-friendly interface and database. We further validate its effectiveness by experimentally identifying anticancer targets of Ginsenoside Rh1. By offering comprehensive data, advanced modeling, and accessible tools, SCOPE-DTI accelerates drug discovery research.
Tokensome: Towards a Genetic Vision-Language GPT for Explainable and Cognitive Karyotyping
Mar 17, 2024



Abstract:Automatic karyotype analysis is often defined as a visual perception task focused solely on chromosomal object-level modeling. This definition has led most existing methods to overlook componential and holistic information, significantly constraining model performance. Moreover, the lack of interpretability in current technologies hinders clinical adoption. In this paper, we introduce Tokensome, a novel vision-language model based on chromosome tokenization for explainable and cognitive karyotyping. Tokensome elevates the method from the conventional visual perception layer to the cognitive decision-making layer. This elevation enables the integration of domain knowledge and cognitive reasoning via knowledge graphs and LLMs, markedly enhancing model's explainability and facilitating abnormality detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge