Xiaoyuan Lu
UE4-NeRF:Neural Radiance Field for Real-Time Rendering of Large-Scale Scene
Oct 20, 2023Abstract:Neural Radiance Fields (NeRF) is a novel implicit 3D reconstruction method that shows immense potential and has been gaining increasing attention. It enables the reconstruction of 3D scenes solely from a set of photographs. However, its real-time rendering capability, especially for interactive real-time rendering of large-scale scenes, still has significant limitations. To address these challenges, in this paper, we propose a novel neural rendering system called UE4-NeRF, specifically designed for real-time rendering of large-scale scenes. We partitioned each large scene into different sub-NeRFs. In order to represent the partitioned independent scene, we initialize polygonal meshes by constructing multiple regular octahedra within the scene and the vertices of the polygonal faces are continuously optimized during the training process. Drawing inspiration from Level of Detail (LOD) techniques, we trained meshes of varying levels of detail for different observation levels. Our approach combines with the rasterization pipeline in Unreal Engine 4 (UE4), achieving real-time rendering of large-scale scenes at 4K resolution with a frame rate of up to 43 FPS. Rendering within UE4 also facilitates scene editing in subsequent stages. Furthermore, through experiments, we have demonstrated that our method achieves rendering quality comparable to state-of-the-art approaches. Project page: https://jamchaos.github.io/UE4-NeRF/.
A Systematic Collection of Medical Image Datasets for Deep Learning
Jun 24, 2021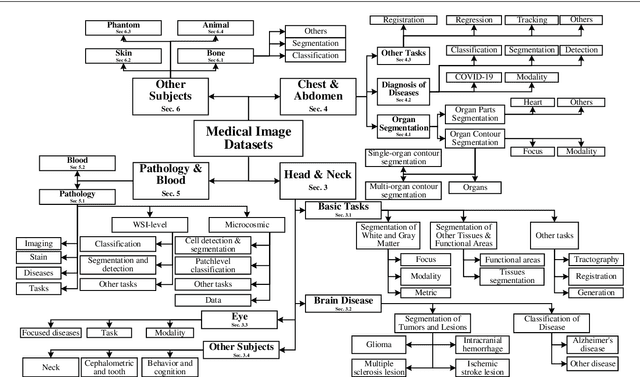
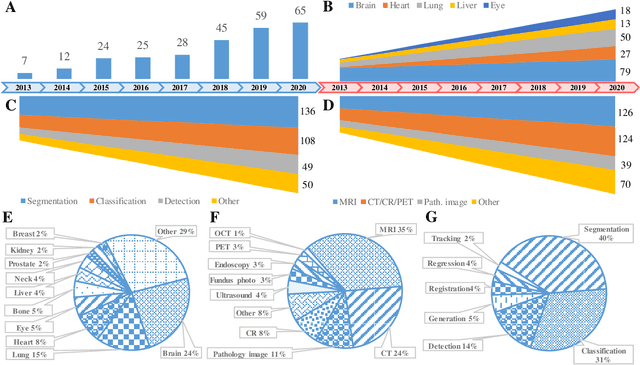
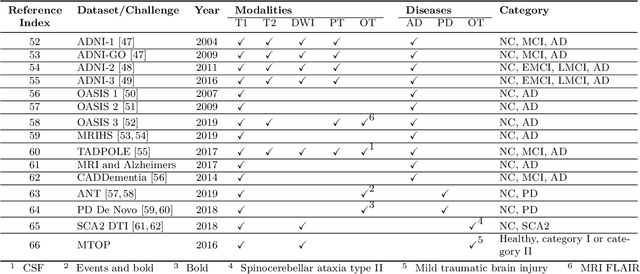
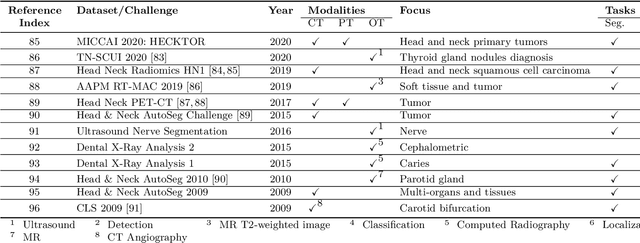
Abstract:The astounding success made by artificial intelligence (AI) in healthcare and other fields proves that AI can achieve human-like performance. However, success always comes with challenges. Deep learning algorithms are data-dependent and require large datasets for training. The lack of data in the medical imaging field creates a bottleneck for the application of deep learning to medical image analysis. Medical image acquisition, annotation, and analysis are costly, and their usage is constrained by ethical restrictions. They also require many resources, such as human expertise and funding. That makes it difficult for non-medical researchers to have access to useful and large medical data. Thus, as comprehensive as possible, this paper provides a collection of medical image datasets with their associated challenges for deep learning research. We have collected information of around three hundred datasets and challenges mainly reported between 2013 and 2020 and categorized them into four categories: head & neck, chest & abdomen, pathology & blood, and ``others''. Our paper has three purposes: 1) to provide a most up to date and complete list that can be used as a universal reference to easily find the datasets for clinical image analysis, 2) to guide researchers on the methodology to test and evaluate their methods' performance and robustness on relevant datasets, 3) to provide a ``route'' to relevant algorithms for the relevant medical topics, and challenge leaderboards.
MeDaS: An open-source platform as service to help break the walls between medicine and informatics
Jul 14, 2020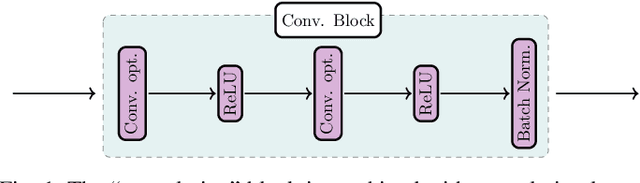

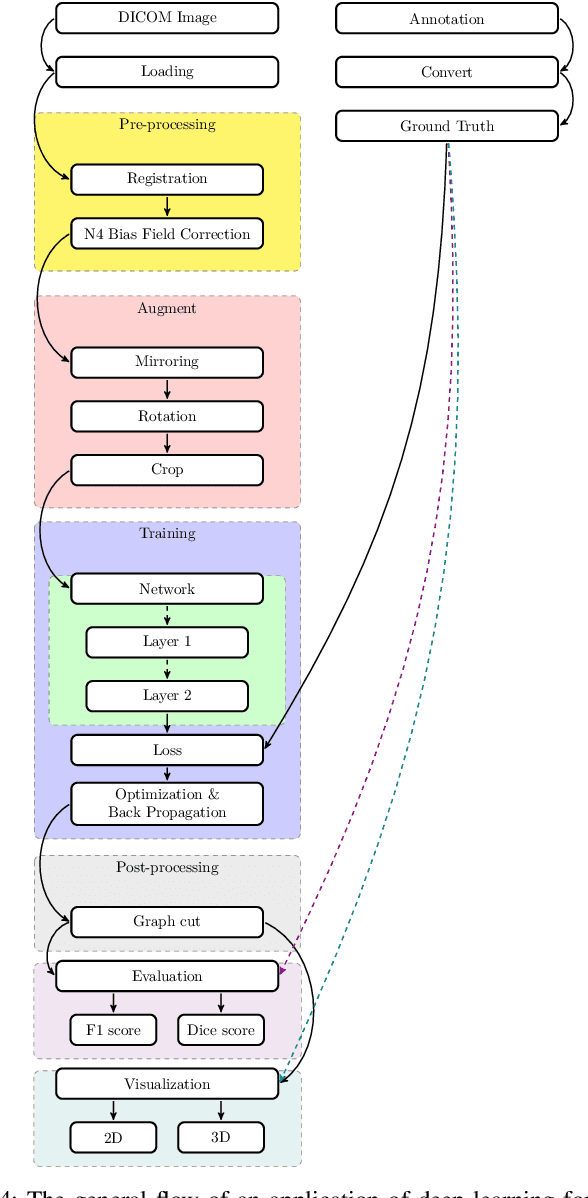
Abstract:In the past decade, deep learning (DL) has achieved unprecedented success in numerous fields including computer vision, natural language processing, and healthcare. In particular, DL is experiencing an increasing development in applications for advanced medical image analysis in terms of analysis, segmentation, classification, and furthermore. On the one hand, tremendous needs that leverage the power of DL for medical image analysis are arising from the research community of a medical, clinical, and informatics background to jointly share their expertise, knowledge, skills, and experience. On the other hand, barriers between disciplines are on the road for them often hampering a full and efficient collaboration. To this end, we propose our novel open-source platform, i.e., MeDaS -- the MeDical open-source platform as Service. To the best of our knowledge, MeDaS is the first open-source platform proving a collaborative and interactive service for researchers from a medical background easily using DL related toolkits, and at the same time for scientists or engineers from information sciences to understand the medical knowledge side. Based on a series of toolkits and utilities from the idea of RINV (Rapid Implementation aNd Verification), our proposed MeDaS platform can implement pre-processing, post-processing, augmentation, visualization, and other phases needed in medical image analysis. Five tasks including the subjects of lung, liver, brain, chest, and pathology, are validated and demonstrated to be efficiently realisable by using MeDaS.
Structure-Feature based Graph Self-adaptive Pooling
Jan 30, 2020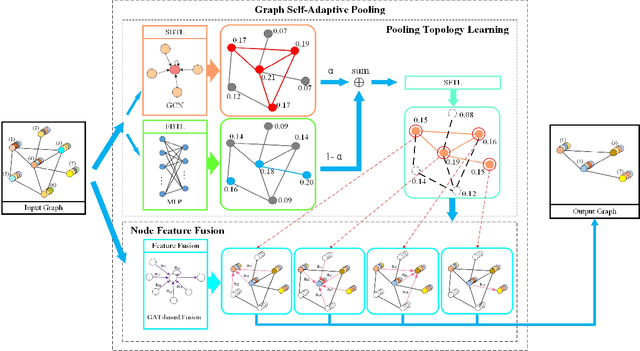

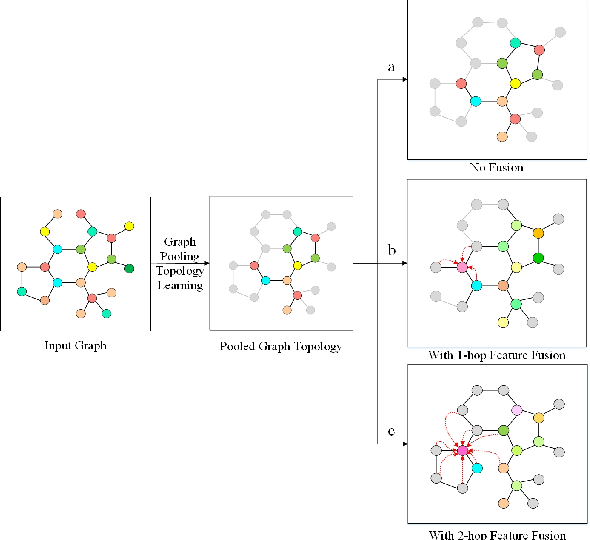
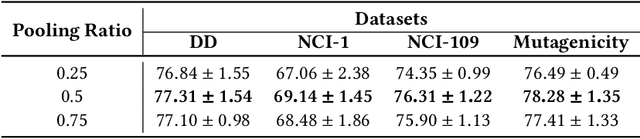
Abstract:Various methods to deal with graph data have been proposed in recent years. However, most of these methods focus on graph feature aggregation rather than graph pooling. Besides, the existing top-k selection graph pooling methods have a few problems. First, to construct the pooled graph topology, current top-k selection methods evaluate the importance of the node from a single perspective only, which is simplistic and unobjective. Second, the feature information of unselected nodes is directly lost during the pooling process, which inevitably leads to a massive loss of graph feature information. To solve these problems mentioned above, we propose a novel graph self-adaptive pooling method with the following objectives: (1) to construct a reasonable pooled graph topology, structure and feature information of the graph are considered simultaneously, which provide additional veracity and objectivity in node selection; and (2) to make the pooled nodes contain sufficiently effective graph information, node feature information is aggregated before discarding the unimportant nodes; thus, the selected nodes contain information from neighbor nodes, which can enhance the use of features of the unselected nodes. Experimental results on four different datasets demonstrate that our method is effective in graph classification and outperforms state-of-the-art graph pooling methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge