Wonsik Jung
IdenBAT: Disentangled Representation Learning for Identity-Preserved Brain Age Transformation
Oct 22, 2024Abstract:Brain age transformation aims to convert reference brain images into synthesized images that accurately reflect the age-specific features of a target age group. The primary objective of this task is to modify only the age-related attributes of the reference image while preserving all other age-irrelevant attributes. However, achieving this goal poses substantial challenges due to the inherent entanglement of various image attributes within features extracted from a backbone encoder, resulting in simultaneous alterations during the image generation. To address this challenge, we propose a novel architecture that employs disentangled representation learning for identity-preserved brain age transformation called IdenBAT. This approach facilitates the decomposition of image features, ensuring the preservation of individual traits while selectively transforming age-related characteristics to match those of the target age group. Through comprehensive experiments conducted on both 2D and full-size 3D brain datasets, our method adeptly converts input images to target age while retaining individual characteristics accurately. Furthermore, our approach demonstrates superiority over existing state-of-the-art regarding performance fidelity.
A Quantitatively Interpretable Model for Alzheimer's Disease Prediction Using Deep Counterfactuals
Oct 05, 2023



Abstract:Deep learning (DL) for predicting Alzheimer's disease (AD) has provided timely intervention in disease progression yet still demands attentive interpretability to explain how their DL models make definitive decisions. Recently, counterfactual reasoning has gained increasing attention in medical research because of its ability to provide a refined visual explanatory map. However, such visual explanatory maps based on visual inspection alone are insufficient unless we intuitively demonstrate their medical or neuroscientific validity via quantitative features. In this study, we synthesize the counterfactual-labeled structural MRIs using our proposed framework and transform it into a gray matter density map to measure its volumetric changes over the parcellated region of interest (ROI). We also devised a lightweight linear classifier to boost the effectiveness of constructed ROIs, promoted quantitative interpretation, and achieved comparable predictive performance to DL methods. Throughout this, our framework produces an ``AD-relatedness index'' for each ROI and offers an intuitive understanding of brain status for an individual patient and across patient groups with respect to AD progression.
Deep Geometric Learning with Monotonicity Constraints for Alzheimer's Disease Progression
Oct 05, 2023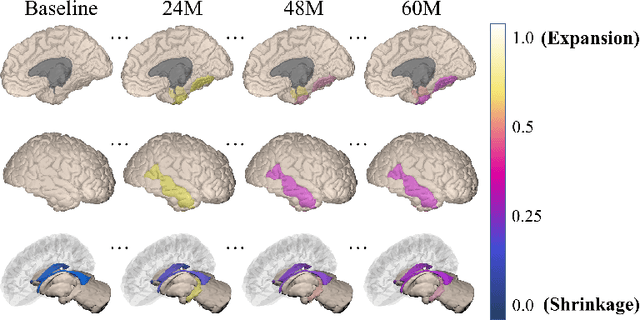
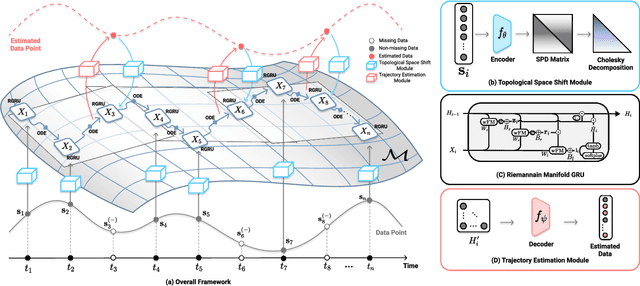
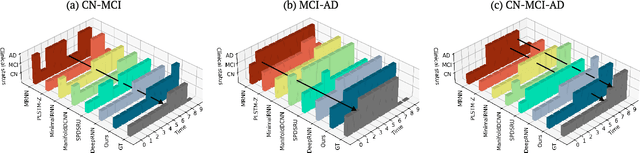
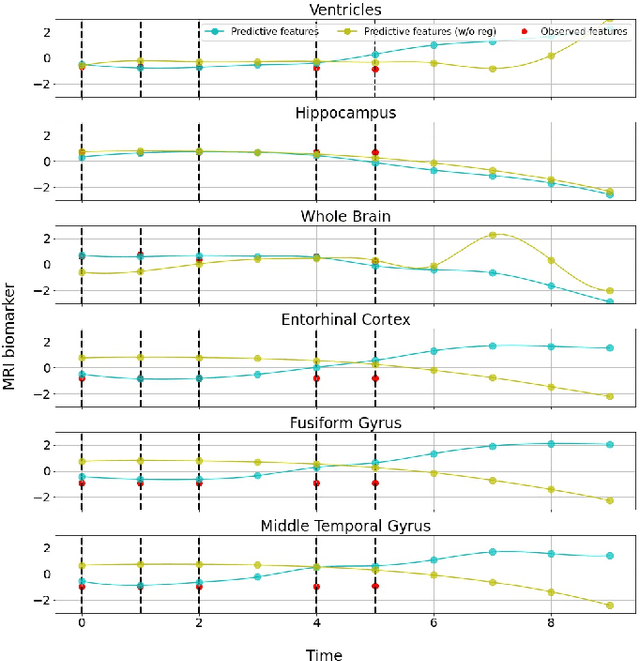
Abstract:Alzheimer's disease (AD) is a devastating neurodegenerative condition that precedes progressive and irreversible dementia; thus, predicting its progression over time is vital for clinical diagnosis and treatment. Numerous studies have implemented structural magnetic resonance imaging (MRI) to model AD progression, focusing on three integral aspects: (i) temporal variability, (ii) incomplete observations, and (iii) temporal geometric characteristics. However, deep learning-based approaches regarding data variability and sparsity have yet to consider inherent geometrical properties sufficiently. The ordinary differential equation-based geometric modeling method (ODE-RGRU) has recently emerged as a promising strategy for modeling time-series data by intertwining a recurrent neural network and an ODE in Riemannian space. Despite its achievements, ODE-RGRU encounters limitations when extrapolating positive definite symmetric metrics from incomplete samples, leading to feature reverse occurrences that are particularly problematic, especially within the clinical facet. Therefore, this study proposes a novel geometric learning approach that models longitudinal MRI biomarkers and cognitive scores by combining three modules: topological space shift, ODE-RGRU, and trajectory estimation. We have also developed a training algorithm that integrates manifold mapping with monotonicity constraints to reflect measurement transition irreversibility. We verify our proposed method's efficacy by predicting clinical labels and cognitive scores over time in regular and irregular settings. Furthermore, we thoroughly analyze our proposed framework through an ablation study.
EAG-RS: A Novel Explainability-guided ROI-Selection Framework for ASD Diagnosis via Inter-regional Relation Learning
Oct 05, 2023



Abstract:Deep learning models based on resting-state functional magnetic resonance imaging (rs-fMRI) have been widely used to diagnose brain diseases, particularly autism spectrum disorder (ASD). Existing studies have leveraged the functional connectivity (FC) of rs-fMRI, achieving notable classification performance. However, they have significant limitations, including the lack of adequate information while using linear low-order FC as inputs to the model, not considering individual characteristics (i.e., different symptoms or varying stages of severity) among patients with ASD, and the non-explainability of the decision process. To cover these limitations, we propose a novel explainability-guided region of interest (ROI) selection (EAG-RS) framework that identifies non-linear high-order functional associations among brain regions by leveraging an explainable artificial intelligence technique and selects class-discriminative regions for brain disease identification. The proposed framework includes three steps: (i) inter-regional relation learning to estimate non-linear relations through random seed-based network masking, (ii) explainable connection-wise relevance score estimation to explore high-order relations between functional connections, and (iii) non-linear high-order FC-based diagnosis-informative ROI selection and classifier learning to identify ASD. We validated the effectiveness of our proposed method by conducting experiments using the Autism Brain Imaging Database Exchange (ABIDE) dataset, demonstrating that the proposed method outperforms other comparative methods in terms of various evaluation metrics. Furthermore, we qualitatively analyzed the selected ROIs and identified ASD subtypes linked to previous neuroscientific studies.
XADLiME: eXplainable Alzheimer's Disease Likelihood Map Estimation via Clinically-guided Prototype Learning
Jul 27, 2022



Abstract:Diagnosing Alzheimer's disease (AD) involves a deliberate diagnostic process owing to its innate traits of irreversibility with subtle and gradual progression. These characteristics make AD biomarker identification from structural brain imaging (e.g., structural MRI) scans quite challenging. Furthermore, there is a high possibility of getting entangled with normal aging. We propose a novel deep-learning approach through eXplainable AD Likelihood Map Estimation (XADLiME) for AD progression modeling over 3D sMRIs using clinically-guided prototype learning. Specifically, we establish a set of topologically-aware prototypes onto the clusters of latent clinical features, uncovering an AD spectrum manifold. We then measure the similarities between latent clinical features and well-established prototypes, estimating a "pseudo" likelihood map. By considering this pseudo map as an enriched reference, we employ an estimating network to estimate the AD likelihood map over a 3D sMRI scan. Additionally, we promote the explainability of such a likelihood map by revealing a comprehensible overview from two perspectives: clinical and morphological. During the inference, this estimated likelihood map served as a substitute over unseen sMRI scans for effectively conducting the downstream task while providing thorough explainable states.
Fine-Grained Attention for Weakly Supervised Object Localization
Apr 11, 2021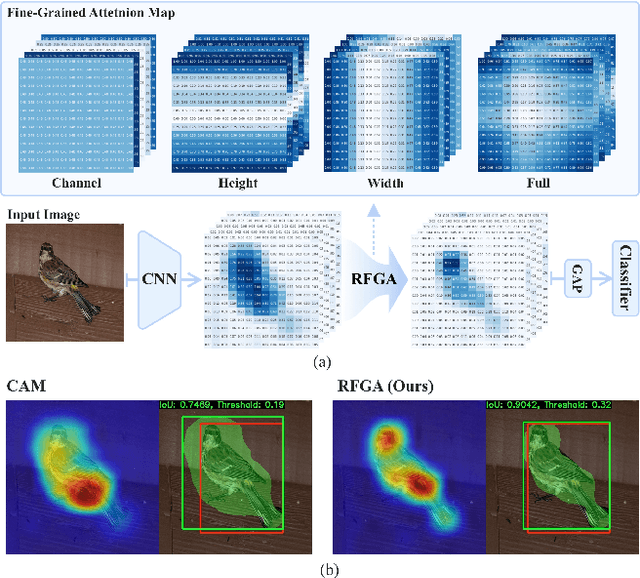
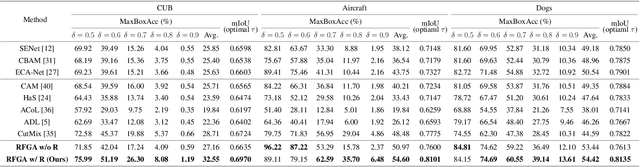
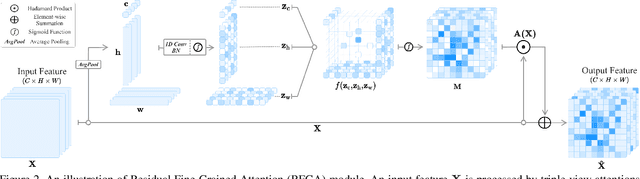
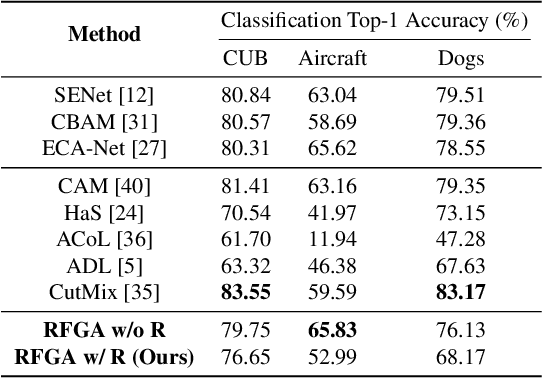
Abstract:Although recent advances in deep learning accelerated an improvement in a weakly supervised object localization (WSOL) task, there are still challenges to identify the entire body of an object, rather than only discriminative parts. In this paper, we propose a novel residual fine-grained attention (RFGA) module that autonomously excites the less activated regions of an object by utilizing information distributed over channels and locations within feature maps in combination with a residual operation. To be specific, we devise a series of mechanisms of triple-view attention representation, attention expansion, and feature calibration. Unlike other attention-based WSOL methods that learn a coarse attention map, having the same values across elements in feature maps, our proposed RFGA learns fine-grained values in an attention map by assigning different attention values for each of the elements. We validated the superiority of our proposed RFGA module by comparing it with the recent methods in the literature over three datasets. Further, we analyzed the effect of each mechanism in our RFGA and visualized attention maps to get insights.
Deep Recurrent Disease Progression Model for Conversion-Time Prediction of Alzheimer's Disease
May 06, 2020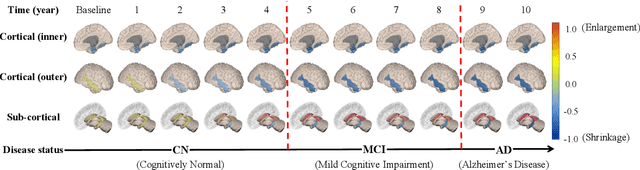
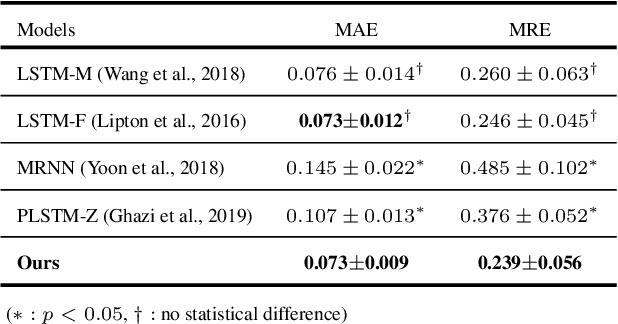
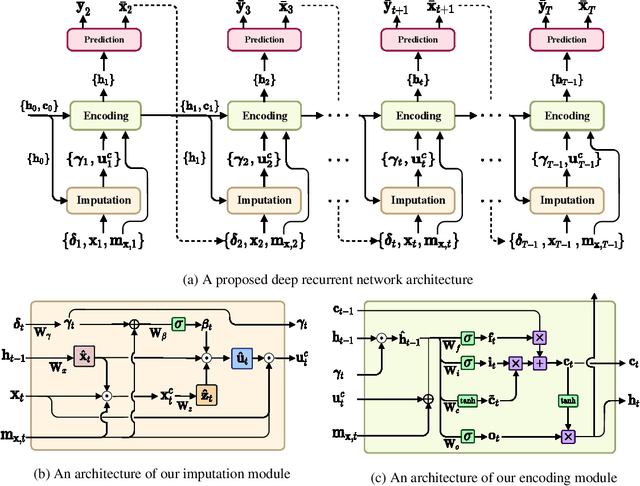
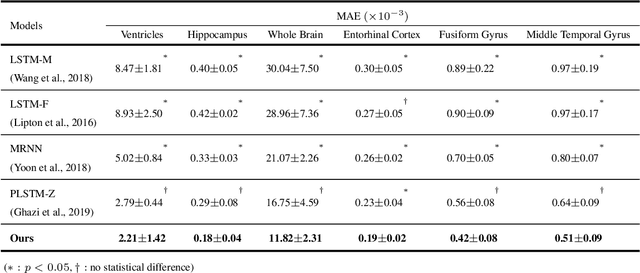
Abstract:Alzheimer's disease (AD) is known as one of the major causes of dementia and is characterized by slow progression over several years, with no treatments or available medicines. In this regard, there have been efforts to identify the risk of developing AD in its earliest time. While many of the previous works considered cross-sectional analysis, more recent studies have focused on the diagnosis and prognosis of AD with longitudinal or time-series data in a way of disease progression modeling (DPM). Under the same problem settings, in this work, we propose a novel computational framework that forecasts the phenotypic measurements of MRI biomarkers and predicts the clinical statuses at multiple future time points. However, in handling time series data, it generally faces with many unexpected missing observations. In regard to such an unfavorable situation, we define a secondary problem of estimating those missing values and tackle it in a systematic way by taking account of temporal and multivariate relations inherent in time series data. Concretely, we propose a deep recurrent network that jointly tackles the three problems of (i) missing value imputation, (ii) phenotypic measurements forecasting, and (iii) clinical status prediction of a subject based on his/her longitudinal imaging biomarkers. Notably, the learnable model parameters of our network are trained in an end to end manner with our circumspectly defined loss function. In our experiments over TADPOLE challenge cohort, we measured performance for various metrics and compared our method to competing methods in the literature. Exhaustive analyses and ablation studies were also conducted to better confirm the effectiveness of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge