Ahmad Wisnu Mulyadi
KindMed: Knowledge-Induced Medicine Prescribing Network for Medication Recommendation
Oct 23, 2023Abstract:Extensive adoption of electronic health records (EHRs) offers opportunities for its use in various clinical analyses. We could acquire more comprehensive insights by enriching an EHR cohort with external knowledge (e.g., standardized medical ontology and wealthy semantics curated on the web) as it divulges a spectrum of informative relations between observed medical codes. This paper proposes a novel Knowledge-Induced Medicine Prescribing Network (KindMed) framework to recommend medicines by inducing knowledge from myriad medical-related external sources upon the EHR cohort, rendering them as medical knowledge graphs (KGs). On top of relation-aware graph representation learning to unravel an adequate embedding of such KGs, we leverage hierarchical sequence learning to discover and fuse clinical and medicine temporal dynamics across patients' historical admissions for encouraging personalized recommendations. In predicting safe, precise, and personalized medicines, we devise an attentive prescribing that accounts for and associates three essential aspects, i.e., a summary of joint historical medical records, clinical condition progression, and the current clinical state of patients. We exhibited the effectiveness of our KindMed on the augmented real-world EHR cohorts, etching leading performances against graph-driven competing baselines.
A Quantitatively Interpretable Model for Alzheimer's Disease Prediction Using Deep Counterfactuals
Oct 05, 2023



Abstract:Deep learning (DL) for predicting Alzheimer's disease (AD) has provided timely intervention in disease progression yet still demands attentive interpretability to explain how their DL models make definitive decisions. Recently, counterfactual reasoning has gained increasing attention in medical research because of its ability to provide a refined visual explanatory map. However, such visual explanatory maps based on visual inspection alone are insufficient unless we intuitively demonstrate their medical or neuroscientific validity via quantitative features. In this study, we synthesize the counterfactual-labeled structural MRIs using our proposed framework and transform it into a gray matter density map to measure its volumetric changes over the parcellated region of interest (ROI). We also devised a lightweight linear classifier to boost the effectiveness of constructed ROIs, promoted quantitative interpretation, and achieved comparable predictive performance to DL methods. Throughout this, our framework produces an ``AD-relatedness index'' for each ROI and offers an intuitive understanding of brain status for an individual patient and across patient groups with respect to AD progression.
XADLiME: eXplainable Alzheimer's Disease Likelihood Map Estimation via Clinically-guided Prototype Learning
Jul 27, 2022



Abstract:Diagnosing Alzheimer's disease (AD) involves a deliberate diagnostic process owing to its innate traits of irreversibility with subtle and gradual progression. These characteristics make AD biomarker identification from structural brain imaging (e.g., structural MRI) scans quite challenging. Furthermore, there is a high possibility of getting entangled with normal aging. We propose a novel deep-learning approach through eXplainable AD Likelihood Map Estimation (XADLiME) for AD progression modeling over 3D sMRIs using clinically-guided prototype learning. Specifically, we establish a set of topologically-aware prototypes onto the clusters of latent clinical features, uncovering an AD spectrum manifold. We then measure the similarities between latent clinical features and well-established prototypes, estimating a "pseudo" likelihood map. By considering this pseudo map as an enriched reference, we employ an estimating network to estimate the AD likelihood map over a 3D sMRI scan. Additionally, we promote the explainability of such a likelihood map by revealing a comprehensible overview from two perspectives: clinical and morphological. During the inference, this estimated likelihood map served as a substitute over unseen sMRI scans for effectively conducting the downstream task while providing thorough explainable states.
Efficient Continuous Manifold Learning for Time Series Modeling
Dec 03, 2021



Abstract:Modeling non-Euclidean data is drawing attention along with the unprecedented successes of deep neural networks in diverse fields. In particular, symmetric positive definite (SPD) matrix is being actively studied in computer vision, signal processing, and medical image analysis, thanks to its ability to learn appropriate statistical representations. However, due to its strong constraints, it remains challenging for optimization problems or inefficient computation costs, especially, within a deep learning framework. In this paper, we propose to exploit a diffeomorphism mapping between Riemannian manifolds and a Cholesky space, by which it becomes feasible not only to efficiently solve optimization problems but also to reduce computation costs greatly. Further, in order for dynamics modeling in time series data, we devise a continuous manifold learning method by integrating a manifold ordinary differential equation and a gated recurrent neural network in a systematic manner. It is noteworthy that because of the nice parameterization of matrices in a Cholesky space, it is straightforward to train our proposed network with Riemannian geometric metrics equipped. We demonstrate through experiments that the proposed model can be efficiently and reliably trained as well as outperform existing manifold methods and state-of-the-art methods in two classification tasks: action recognition and sleep staging classification.
Uncertainty-Aware Variational-Recurrent Imputation Network for Clinical Time Series
Mar 02, 2020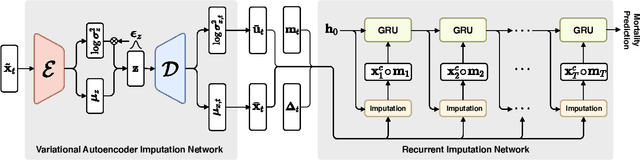
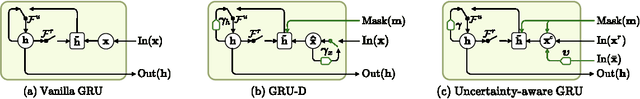
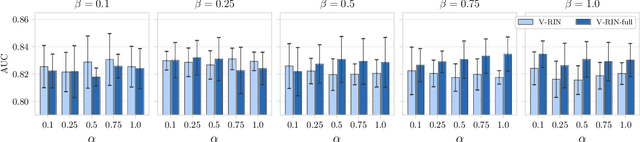
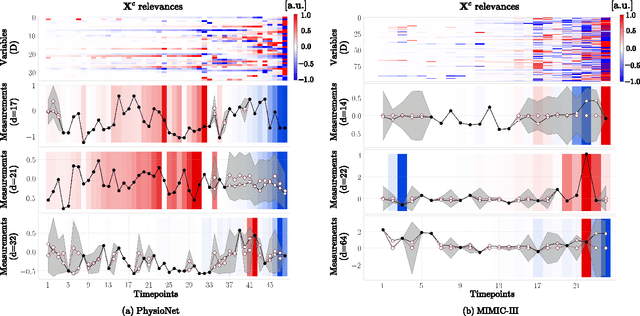
Abstract:Electronic health records (EHR) consist of longitudinal clinical observations portrayed with sparsity, irregularity, and high-dimensionality, which become major obstacles in drawing reliable downstream clinical outcomes. Although there exist great numbers of imputation methods to tackle these issues, most of them ignore correlated features, temporal dynamics and entirely set aside the uncertainty. Since the missing value estimates involve the risk of being inaccurate, it is appropriate for the method to handle the less certain information differently than the reliable data. In that regard, we can use the uncertainties in estimating the missing values as the fidelity score to be further utilized to alleviate the risk of biased missing value estimates. In this work, we propose a novel variational-recurrent imputation network, which unifies an imputation and a prediction network by taking into account the correlated features, temporal dynamics, as well as the uncertainty. Specifically, we leverage the deep generative model in the imputation, which is based on the distribution among variables, and a recurrent imputation network to exploit the temporal relations, in conjunction with utilization of the uncertainty. We validated the effectiveness of our proposed model on two publicly available real-world EHR datasets: PhysioNet Challenge 2012 and MIMIC-III, and compared the results with other competing state-of-the-art methods in the literature.
Uncertainty-Gated Stochastic Sequential Model for EHR Mortality Prediction
Mar 02, 2020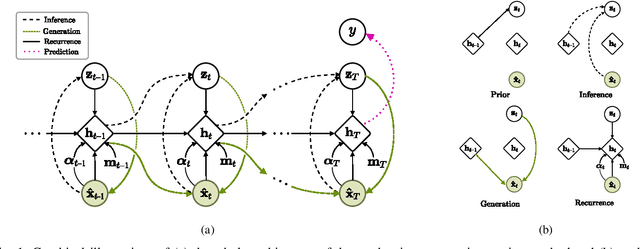
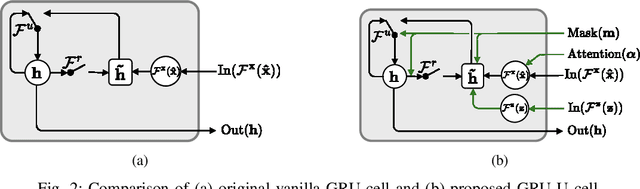
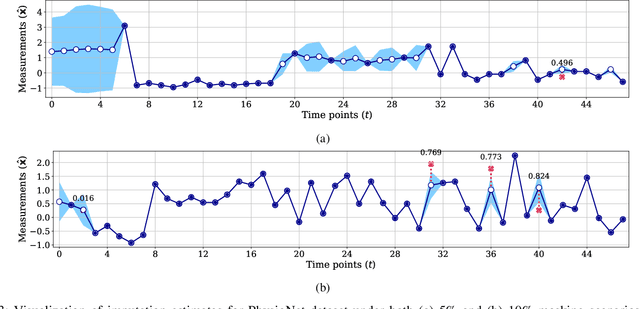
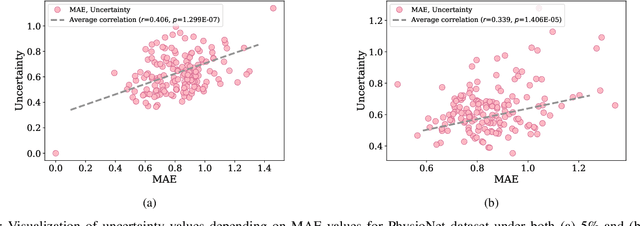
Abstract:Electronic health records (EHR) are characterized as non-stationary, heterogeneous, noisy, and sparse data; therefore, it is challenging to learn the regularities or patterns inherent within them. In particular, sparseness caused mostly by many missing values has attracted the attention of researchers, who have attempted to find a better use of all available samples for determining the solution of a primary target task through the defining a secondary imputation problem. Methodologically, existing methods, either deterministic or stochastic, have applied different assumptions to impute missing values. However, once the missing values are imputed, most existing methods do not consider the fidelity or confidence of the imputed values in the modeling of downstream tasks. Undoubtedly, an erroneous or improper imputation of missing variables can cause difficulties in modeling as well as a degraded performance. In this study, we present a novel variational recurrent network that (i) estimates the distribution of missing variables allowing to represent uncertainty in the imputed values, (ii) updates hidden states by explicitly applying fidelity based on a variance of the imputed values during a recurrence (i.e., uncertainty propagation over time), and (iii) predicts the possibility of in-hospital mortality. It is noteworthy that our model can conduct these procedures in a single stream and learn all network parameters jointly in an end-to-end manner. We validated the effectiveness of our method using the public datasets of MIMIC-III and PhysioNet challenge 2012 by comparing with and outperforming other state-of-the-art methods for mortality prediction considered in our experiments. In addition, we identified the behavior of the model that well represented the uncertainties for the imputed estimates, which indicated a high correlation between the calculated MAE and the uncertainty.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge