Eunji Jun
Medical Transformer: Universal Brain Encoder for 3D MRI Analysis
Apr 28, 2021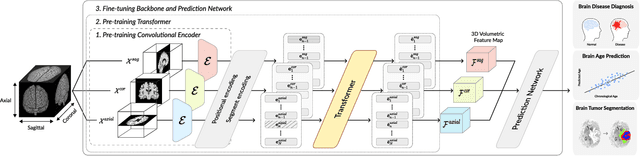
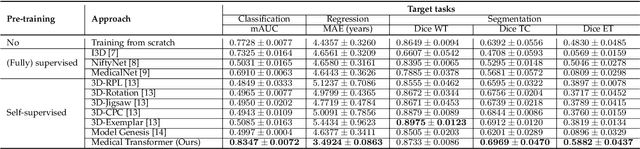
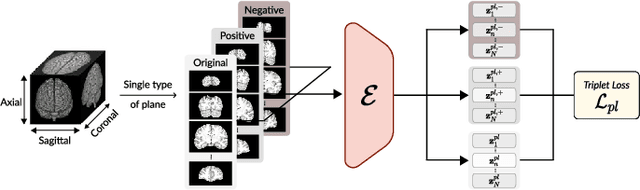
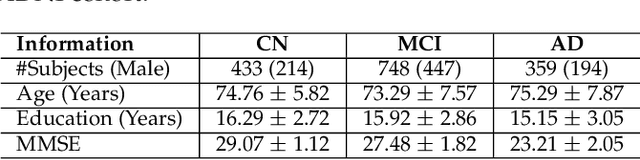
Abstract:Transfer learning has gained attention in medical image analysis due to limited annotated 3D medical datasets for training data-driven deep learning models in the real world. Existing 3D-based methods have transferred the pre-trained models to downstream tasks, which achieved promising results with only a small number of training samples. However, they demand a massive amount of parameters to train the model for 3D medical imaging. In this work, we propose a novel transfer learning framework, called Medical Transformer, that effectively models 3D volumetric images in the form of a sequence of 2D image slices. To make a high-level representation in 3D-form empowering spatial relations better, we take a multi-view approach that leverages plenty of information from the three planes of 3D volume, while providing parameter-efficient training. For building a source model generally applicable to various tasks, we pre-train the model in a self-supervised learning manner for masked encoding vector prediction as a proxy task, using a large-scale normal, healthy brain magnetic resonance imaging (MRI) dataset. Our pre-trained model is evaluated on three downstream tasks: (i) brain disease diagnosis, (ii) brain age prediction, and (iii) brain tumor segmentation, which are actively studied in brain MRI research. The experimental results show that our Medical Transformer outperforms the state-of-the-art transfer learning methods, efficiently reducing the number of parameters up to about 92% for classification and
Multi-view Integration Learning for Irregularly-sampled Clinical Time Series
Jan 26, 2021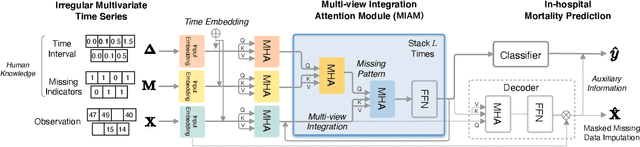
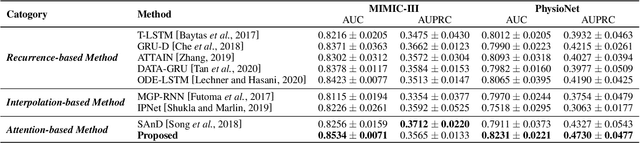
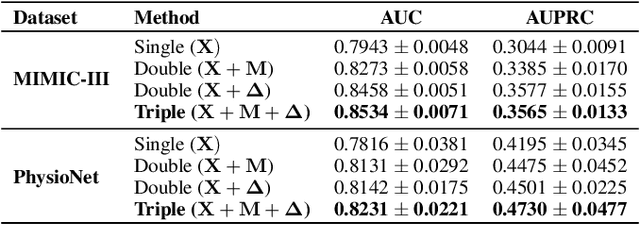
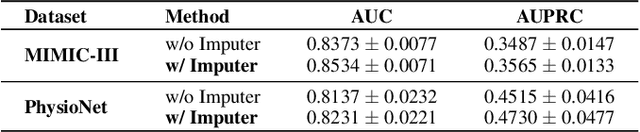
Abstract:Electronic health record (EHR) data is sparse and irregular as it is recorded at irregular time intervals, and different clinical variables are measured at each observation point. In this work, we propose a multi-view features integration learning from irregular multivariate time series data by self-attention mechanism in an imputation-free manner. Specifically, we devise a novel multi-integration attention module (MIAM) to extract complex information inherent in irregular time series data. In particular, we explicitly learn the relationships among the observed values, missing indicators, and time interval between the consecutive observations, simultaneously. The rationale behind our approach is the use of human knowledge such as what to measure and when to measure in different situations, which are indirectly represented in the data. In addition, we build an attention-based decoder as a missing value imputer that helps empower the representation learning of the inter-relations among multi-view observations for the prediction task, which operates at the training phase only. We validated the effectiveness of our method over the public MIMIC-III and PhysioNet challenge 2012 datasets by comparing with and outperforming the state-of-the-art methods for in-hospital mortality prediction.
Deep Recurrent Disease Progression Model for Conversion-Time Prediction of Alzheimer's Disease
May 06, 2020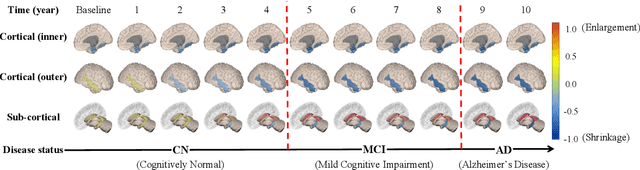
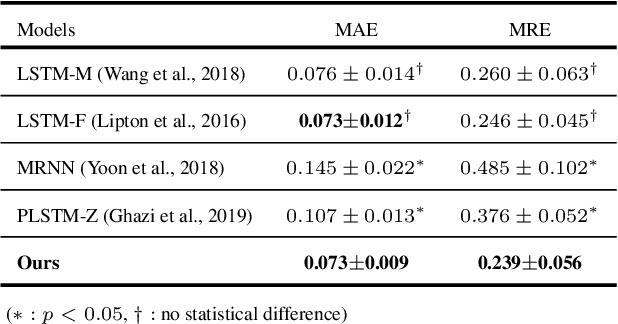
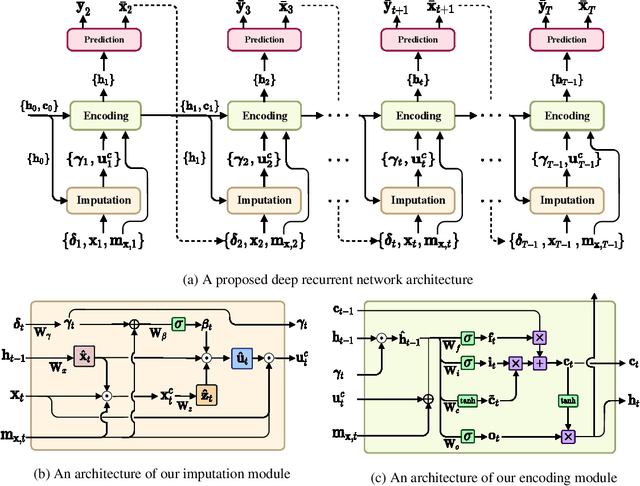
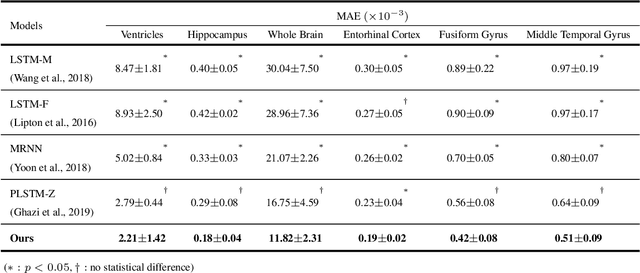
Abstract:Alzheimer's disease (AD) is known as one of the major causes of dementia and is characterized by slow progression over several years, with no treatments or available medicines. In this regard, there have been efforts to identify the risk of developing AD in its earliest time. While many of the previous works considered cross-sectional analysis, more recent studies have focused on the diagnosis and prognosis of AD with longitudinal or time-series data in a way of disease progression modeling (DPM). Under the same problem settings, in this work, we propose a novel computational framework that forecasts the phenotypic measurements of MRI biomarkers and predicts the clinical statuses at multiple future time points. However, in handling time series data, it generally faces with many unexpected missing observations. In regard to such an unfavorable situation, we define a secondary problem of estimating those missing values and tackle it in a systematic way by taking account of temporal and multivariate relations inherent in time series data. Concretely, we propose a deep recurrent network that jointly tackles the three problems of (i) missing value imputation, (ii) phenotypic measurements forecasting, and (iii) clinical status prediction of a subject based on his/her longitudinal imaging biomarkers. Notably, the learnable model parameters of our network are trained in an end to end manner with our circumspectly defined loss function. In our experiments over TADPOLE challenge cohort, we measured performance for various metrics and compared our method to competing methods in the literature. Exhaustive analyses and ablation studies were also conducted to better confirm the effectiveness of our method.
Uncertainty-Aware Variational-Recurrent Imputation Network for Clinical Time Series
Mar 02, 2020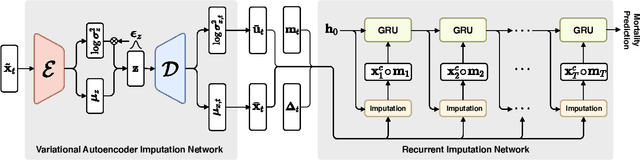
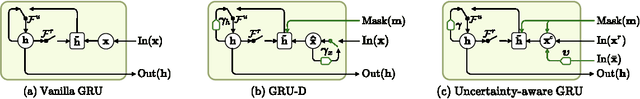
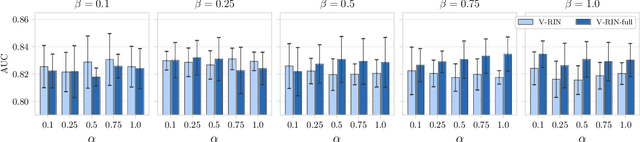
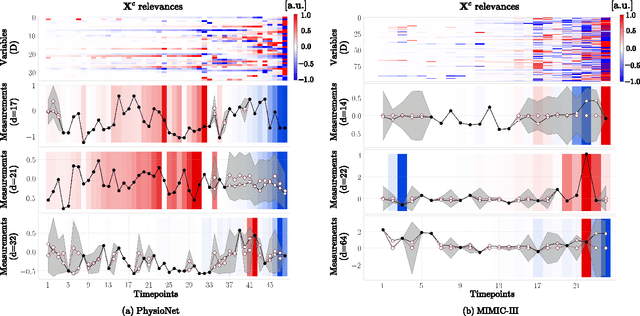
Abstract:Electronic health records (EHR) consist of longitudinal clinical observations portrayed with sparsity, irregularity, and high-dimensionality, which become major obstacles in drawing reliable downstream clinical outcomes. Although there exist great numbers of imputation methods to tackle these issues, most of them ignore correlated features, temporal dynamics and entirely set aside the uncertainty. Since the missing value estimates involve the risk of being inaccurate, it is appropriate for the method to handle the less certain information differently than the reliable data. In that regard, we can use the uncertainties in estimating the missing values as the fidelity score to be further utilized to alleviate the risk of biased missing value estimates. In this work, we propose a novel variational-recurrent imputation network, which unifies an imputation and a prediction network by taking into account the correlated features, temporal dynamics, as well as the uncertainty. Specifically, we leverage the deep generative model in the imputation, which is based on the distribution among variables, and a recurrent imputation network to exploit the temporal relations, in conjunction with utilization of the uncertainty. We validated the effectiveness of our proposed model on two publicly available real-world EHR datasets: PhysioNet Challenge 2012 and MIMIC-III, and compared the results with other competing state-of-the-art methods in the literature.
Uncertainty-Gated Stochastic Sequential Model for EHR Mortality Prediction
Mar 02, 2020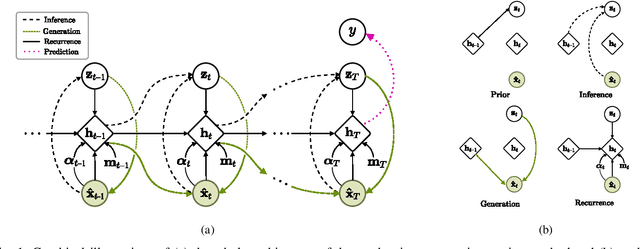
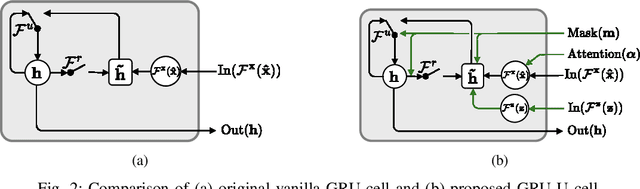
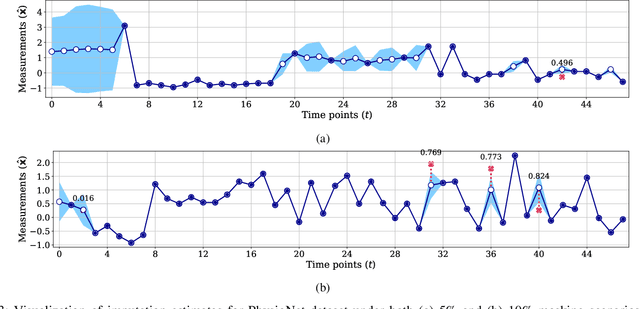
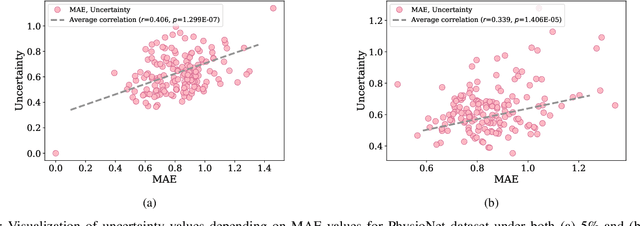
Abstract:Electronic health records (EHR) are characterized as non-stationary, heterogeneous, noisy, and sparse data; therefore, it is challenging to learn the regularities or patterns inherent within them. In particular, sparseness caused mostly by many missing values has attracted the attention of researchers, who have attempted to find a better use of all available samples for determining the solution of a primary target task through the defining a secondary imputation problem. Methodologically, existing methods, either deterministic or stochastic, have applied different assumptions to impute missing values. However, once the missing values are imputed, most existing methods do not consider the fidelity or confidence of the imputed values in the modeling of downstream tasks. Undoubtedly, an erroneous or improper imputation of missing variables can cause difficulties in modeling as well as a degraded performance. In this study, we present a novel variational recurrent network that (i) estimates the distribution of missing variables allowing to represent uncertainty in the imputed values, (ii) updates hidden states by explicitly applying fidelity based on a variance of the imputed values during a recurrence (i.e., uncertainty propagation over time), and (iii) predicts the possibility of in-hospital mortality. It is noteworthy that our model can conduct these procedures in a single stream and learn all network parameters jointly in an end-to-end manner. We validated the effectiveness of our method using the public datasets of MIMIC-III and PhysioNet challenge 2012 by comparing with and outperforming other state-of-the-art methods for mortality prediction considered in our experiments. In addition, we identified the behavior of the model that well represented the uncertainties for the imputed estimates, which indicated a high correlation between the calculated MAE and the uncertainty.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge