Wonjun Ko
TransSleep: Transitioning-aware Attention-based Deep Neural Network for Sleep Staging
Mar 22, 2022
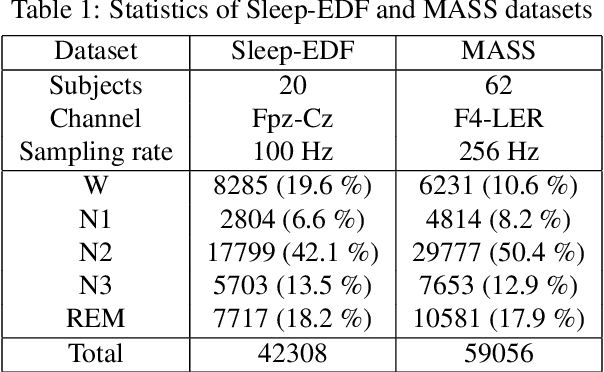

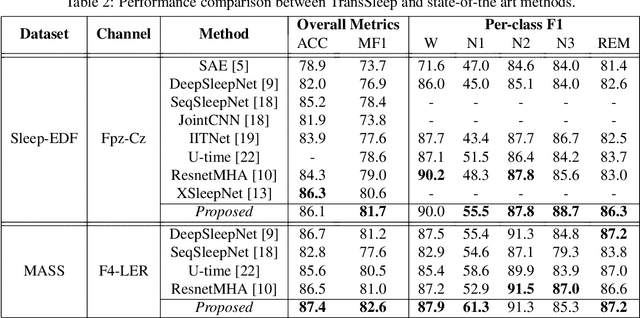
Abstract:Sleep staging is essential for sleep assessment and plays a vital role as a health indicator. Many recent studies have devised various machine learning as well as deep learning architectures for sleep staging. However, two key challenges hinder the practical use of these architectures: effectively capturing salient waveforms in sleep signals and correctly classifying confusing stages in transitioning epochs. In this study, we propose a novel deep neural network structure, TransSleep, that captures distinctive local temporal patterns and distinguishes confusing stages using two auxiliary tasks. In particular, TransSleep adopts an attention-based multi-scale feature extractor module to capture salient waveforms; a stage-confusion estimator module with a novel auxiliary task, epoch-level stage classification, to estimate confidence scores for identifying confusing stages; and a context encoder module with the other novel auxiliary task, stage-transition detection, to represent contextual relationships across neighboring epochs. Results show that TransSleep achieves promising performance in automatic sleep staging. The validity of TransSleep is demonstrated by its state-of-the-art performance on two publicly available datasets, Sleep-EDF and MASS. Furthermore, we performed ablations to analyze our results from different perspectives. Based on our overall results, we believe that TransSleep has immense potential to provide new insights into deep learning-based sleep staging.
Efficient Continuous Manifold Learning for Time Series Modeling
Dec 03, 2021



Abstract:Modeling non-Euclidean data is drawing attention along with the unprecedented successes of deep neural networks in diverse fields. In particular, symmetric positive definite (SPD) matrix is being actively studied in computer vision, signal processing, and medical image analysis, thanks to its ability to learn appropriate statistical representations. However, due to its strong constraints, it remains challenging for optimization problems or inefficient computation costs, especially, within a deep learning framework. In this paper, we propose to exploit a diffeomorphism mapping between Riemannian manifolds and a Cholesky space, by which it becomes feasible not only to efficiently solve optimization problems but also to reduce computation costs greatly. Further, in order for dynamics modeling in time series data, we devise a continuous manifold learning method by integrating a manifold ordinary differential equation and a gated recurrent neural network in a systematic manner. It is noteworthy that because of the nice parameterization of matrices in a Cholesky space, it is straightforward to train our proposed network with Riemannian geometric metrics equipped. We demonstrate through experiments that the proposed model can be efficiently and reliably trained as well as outperform existing manifold methods and state-of-the-art methods in two classification tasks: action recognition and sleep staging classification.
A Novel RL-assisted Deep Learning Framework for Task-informative Signals Selection and Classification for Spontaneous BCIs
Jul 01, 2020


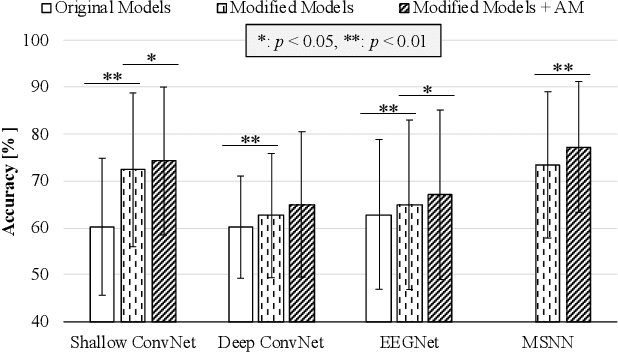
Abstract:In this work, we formulate the problem of estimating and selecting task-relevant temporal signal segments from a single EEG trial in the form of a Markov decision process and propose a novel reinforcement-learning mechanism that can be combined with the existing deep-learning based BCI methods. To be specific, we devise an actor-critic network such that an agent can determine which timepoints need to be used (informative) or discarded (uninformative) in composing the intention-related features in a given trial, and thus enhancing the intention identification performance. To validate the effectiveness of our proposed method, we conducted experiments with a publicly available big MI dataset and applied our novel mechanism to various recent deep-learning architectures designed for MI classification. Based on the exhaustive experiments, we observed that our proposed method helped achieve statistically significant improvements in performance.
Multi-Scale Neural network for EEG Representation Learning in BCI
Mar 02, 2020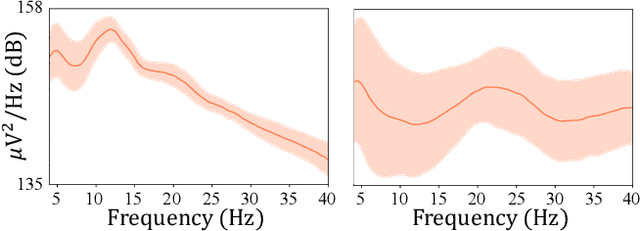


Abstract:Recent advances in deep learning have had a methodological and practical impact on brain-computer interface research. Among the various deep network architectures, convolutional neural networks have been well suited for spatio-spectral-temporal electroencephalogram signal representation learning. Most of the existing CNN-based methods described in the literature extract features at a sequential level of abstraction with repetitive nonlinear operations and involve densely connected layers for classification. However, studies in neurophysiology have revealed that EEG signals carry information in different ranges of frequency components. To better reflect these multi-frequency properties in EEGs, we propose a novel deep multi-scale neural network that discovers feature representations in multiple frequency/time ranges and extracts relationships among electrodes, i.e., spatial representations, for subject intention/condition identification. Furthermore, by completely representing EEG signals with spatio-spectral-temporal information, the proposed method can be utilized for diverse paradigms in both active and passive BCIs, contrary to existing methods that are primarily focused on single-paradigm BCIs. To demonstrate the validity of our proposed method, we conducted experiments on various paradigms of active/passive BCI datasets. Our experimental results demonstrated that the proposed method achieved performance improvements when judged against comparable state-of-the-art methods. Additionally, we analyzed the proposed method using different techniques, such as PSD curves and relevance score inspection to validate the multi-scale EEG signal information capturing ability, activation pattern maps for investigating the learned spatial filters, and t-SNE plotting for visualizing represented features. Finally, we also demonstrated our method's application to real-world problems.
Toward Subject Invariant and Class Disentangled Representation in BCI via Cross-Domain Mutual Information Estimator
Oct 18, 2019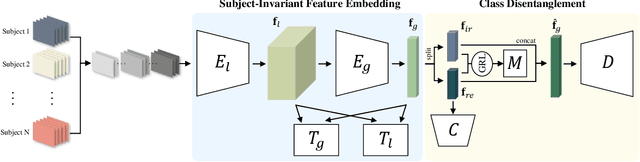
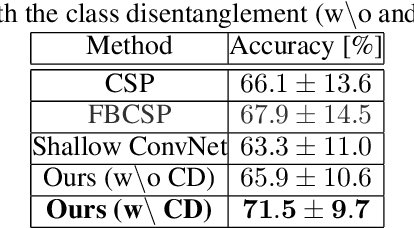
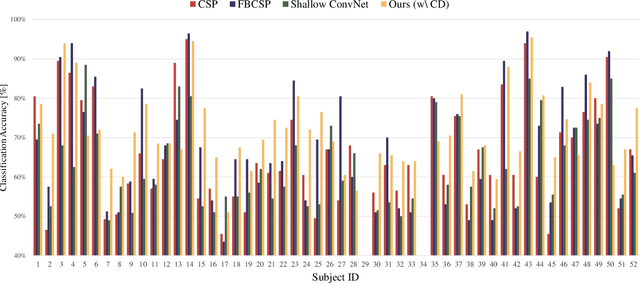

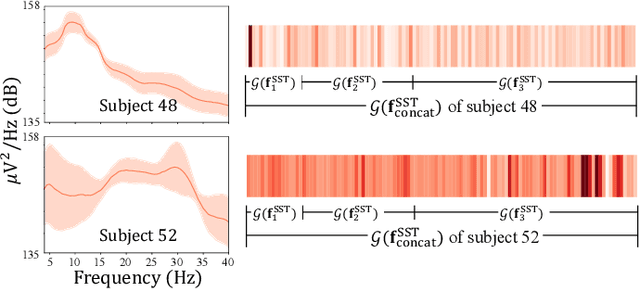
Abstract:In recent, deep learning-based feature representation methods have shown a promising impact in electroencephalography (EEG)-based brain-computer interface (BCI). Nonetheless, due to high intra- and inter-subject variabilities, many studies on decoding EEG were designed in a subject-specific manner by using calibration samples, with no much concern on its less practical, time-consuming, and data-hungry process. To tackle this problem, recent studies took advantage of transfer learning, especially using domain adaptation techniques. However, there still remain two challenging limitations; i) most domain adaptation methods are designed for labeled source and unlabeled target domain whereas BCI tasks generally have multiple annotated domains. ii) most of the methods do not consider negatively transferable to disrupt generalization ability. In this paper, we propose a novel network architecture to tackle those limitations by estimating mutual information in high-level representation and low-level representation, separately. Specifically, our proposed method extracts domain-invariant and class-relevant features, thereby enhancing generalizability in classification across. It is also noteworthy that our method can be applicable to a new subject with a small amount of data via a fine-tuning, step only, reducing calibration time for practical uses. We validated our proposed method on a big motor imagery EEG dataset by showing promising results, compared to competing methods considered in our experiments.
Plug-in Factorization for Latent Representation Disentanglement
May 29, 2019



Abstract:In this work, we propose a Factorized Disentangler-Entangler Network (FDEN) that learns to decompose a latent representation into two mutually independent factors, namely, identity and style. Given a latent representation, the proposed framework draws a set of interpretable factors aligned to identity of an observed data and learns to maximize the independency between these factors. Our work introduces an idea for a plug-in method to disentangle latent representations of already learned deep models with no affect to the model. In doing so, it brings the possibilities of extending state-of-the-art models to solve different tasks and also maintain the performance of its original task. Thus, FDEN is naturally applicable to jointly perform multiple tasks such as few-shot learning and image-to-image translation in a single framework. We show the effectiveness of our work in disentangling a latent representation in two parts. First, to evaluate the alignment of factor to an identity, we perform few-shot learning using only the aligned factor. Then, to evaluate the effectiveness of decomposition of latent representation and to show that plugin method does not affect the deep model in its performance, we perform image-to-image style transfer by mixing factors of different images. These evaluations show, qualitatively and quantitatively, that our proposed framework can indeed disentangle a latent representation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge