Eunjin Jeon
FIESTA: Fourier-Based Semantic Augmentation with Uncertainty Guidance for Enhanced Domain Generalizability in Medical Image Segmentation
Jun 20, 2024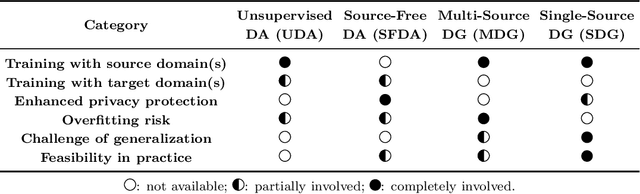
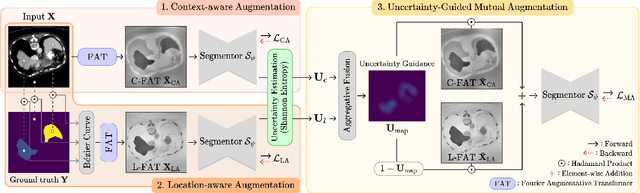
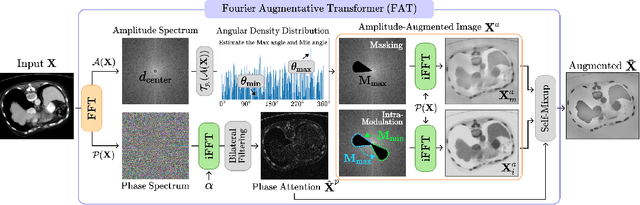
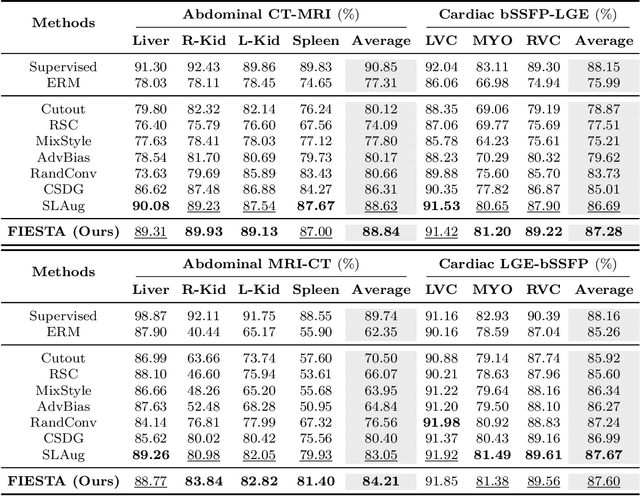
Abstract:Single-source domain generalization (SDG) in medical image segmentation (MIS) aims to generalize a model using data from only one source domain to segment data from an unseen target domain. Despite substantial advances in SDG with data augmentation, existing methods often fail to fully consider the details and uncertain areas prevalent in MIS, leading to mis-segmentation. This paper proposes a Fourier-based semantic augmentation method called FIESTA using uncertainty guidance to enhance the fundamental goals of MIS in an SDG context by manipulating the amplitude and phase components in the frequency domain. The proposed Fourier augmentative transformer addresses semantic amplitude modulation based on meaningful angular points to induce pertinent variations and harnesses the phase spectrum to ensure structural coherence. Moreover, FIESTA employs epistemic uncertainty to fine-tune the augmentation process, improving the ability of the model to adapt to diverse augmented data and concentrate on areas with higher ambiguity. Extensive experiments across three cross-domain scenarios demonstrate that FIESTA surpasses recent state-of-the-art SDG approaches in segmentation performance and significantly contributes to boosting the applicability of the model in medical imaging modalities.
EAG-RS: A Novel Explainability-guided ROI-Selection Framework for ASD Diagnosis via Inter-regional Relation Learning
Oct 05, 2023



Abstract:Deep learning models based on resting-state functional magnetic resonance imaging (rs-fMRI) have been widely used to diagnose brain diseases, particularly autism spectrum disorder (ASD). Existing studies have leveraged the functional connectivity (FC) of rs-fMRI, achieving notable classification performance. However, they have significant limitations, including the lack of adequate information while using linear low-order FC as inputs to the model, not considering individual characteristics (i.e., different symptoms or varying stages of severity) among patients with ASD, and the non-explainability of the decision process. To cover these limitations, we propose a novel explainability-guided region of interest (ROI) selection (EAG-RS) framework that identifies non-linear high-order functional associations among brain regions by leveraging an explainable artificial intelligence technique and selects class-discriminative regions for brain disease identification. The proposed framework includes three steps: (i) inter-regional relation learning to estimate non-linear relations through random seed-based network masking, (ii) explainable connection-wise relevance score estimation to explore high-order relations between functional connections, and (iii) non-linear high-order FC-based diagnosis-informative ROI selection and classifier learning to identify ASD. We validated the effectiveness of our proposed method by conducting experiments using the Autism Brain Imaging Database Exchange (ABIDE) dataset, demonstrating that the proposed method outperforms other comparative methods in terms of various evaluation metrics. Furthermore, we qualitatively analyzed the selected ROIs and identified ASD subtypes linked to previous neuroscientific studies.
TransSleep: Transitioning-aware Attention-based Deep Neural Network for Sleep Staging
Mar 22, 2022
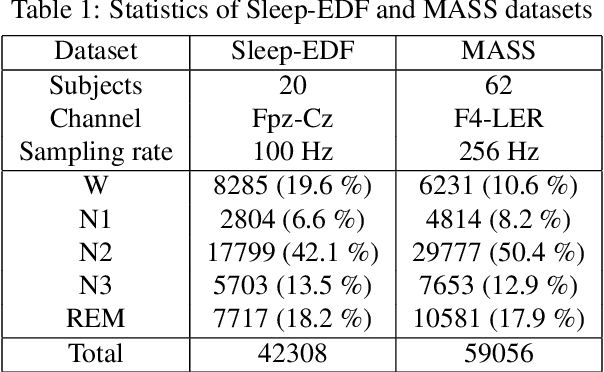

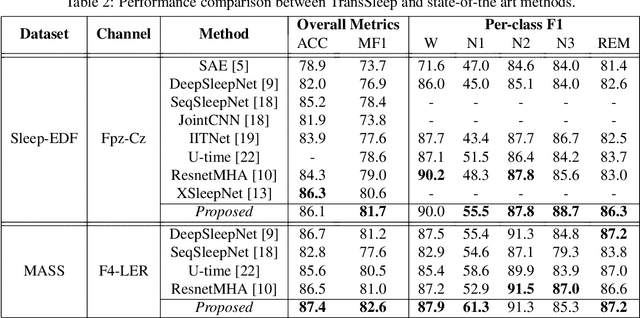
Abstract:Sleep staging is essential for sleep assessment and plays a vital role as a health indicator. Many recent studies have devised various machine learning as well as deep learning architectures for sleep staging. However, two key challenges hinder the practical use of these architectures: effectively capturing salient waveforms in sleep signals and correctly classifying confusing stages in transitioning epochs. In this study, we propose a novel deep neural network structure, TransSleep, that captures distinctive local temporal patterns and distinguishes confusing stages using two auxiliary tasks. In particular, TransSleep adopts an attention-based multi-scale feature extractor module to capture salient waveforms; a stage-confusion estimator module with a novel auxiliary task, epoch-level stage classification, to estimate confidence scores for identifying confusing stages; and a context encoder module with the other novel auxiliary task, stage-transition detection, to represent contextual relationships across neighboring epochs. Results show that TransSleep achieves promising performance in automatic sleep staging. The validity of TransSleep is demonstrated by its state-of-the-art performance on two publicly available datasets, Sleep-EDF and MASS. Furthermore, we performed ablations to analyze our results from different perspectives. Based on our overall results, we believe that TransSleep has immense potential to provide new insights into deep learning-based sleep staging.
Fine-Grained Attention for Weakly Supervised Object Localization
Apr 11, 2021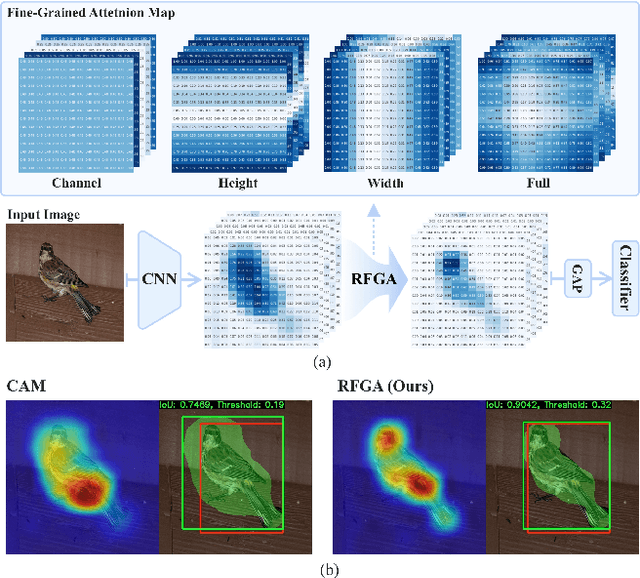
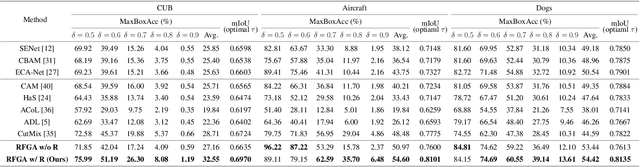
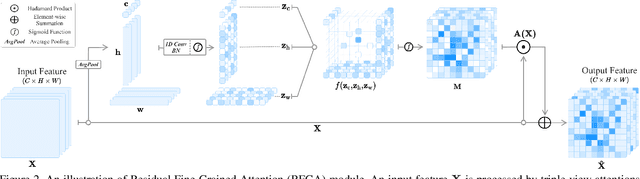
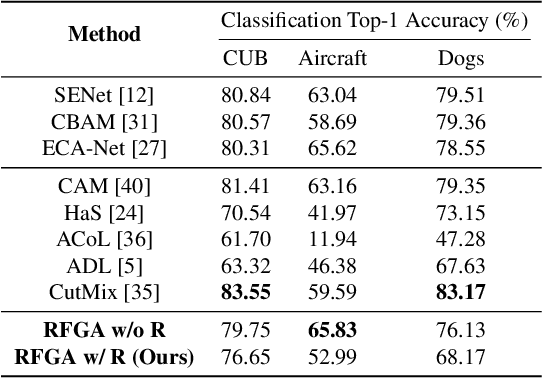
Abstract:Although recent advances in deep learning accelerated an improvement in a weakly supervised object localization (WSOL) task, there are still challenges to identify the entire body of an object, rather than only discriminative parts. In this paper, we propose a novel residual fine-grained attention (RFGA) module that autonomously excites the less activated regions of an object by utilizing information distributed over channels and locations within feature maps in combination with a residual operation. To be specific, we devise a series of mechanisms of triple-view attention representation, attention expansion, and feature calibration. Unlike other attention-based WSOL methods that learn a coarse attention map, having the same values across elements in feature maps, our proposed RFGA learns fine-grained values in an attention map by assigning different attention values for each of the elements. We validated the superiority of our proposed RFGA module by comparing it with the recent methods in the literature over three datasets. Further, we analyzed the effect of each mechanism in our RFGA and visualized attention maps to get insights.
A Novel RL-assisted Deep Learning Framework for Task-informative Signals Selection and Classification for Spontaneous BCIs
Jul 01, 2020


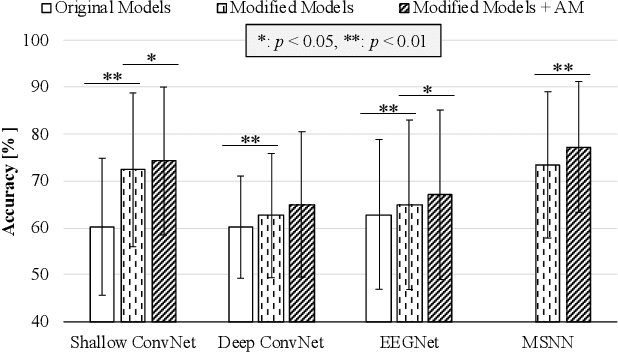
Abstract:In this work, we formulate the problem of estimating and selecting task-relevant temporal signal segments from a single EEG trial in the form of a Markov decision process and propose a novel reinforcement-learning mechanism that can be combined with the existing deep-learning based BCI methods. To be specific, we devise an actor-critic network such that an agent can determine which timepoints need to be used (informative) or discarded (uninformative) in composing the intention-related features in a given trial, and thus enhancing the intention identification performance. To validate the effectiveness of our proposed method, we conducted experiments with a publicly available big MI dataset and applied our novel mechanism to various recent deep-learning architectures designed for MI classification. Based on the exhaustive experiments, we observed that our proposed method helped achieve statistically significant improvements in performance.
Multi-Scale Neural network for EEG Representation Learning in BCI
Mar 02, 2020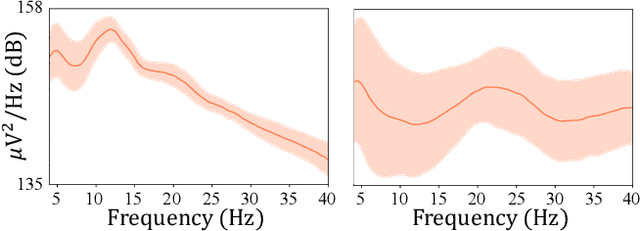


Abstract:Recent advances in deep learning have had a methodological and practical impact on brain-computer interface research. Among the various deep network architectures, convolutional neural networks have been well suited for spatio-spectral-temporal electroencephalogram signal representation learning. Most of the existing CNN-based methods described in the literature extract features at a sequential level of abstraction with repetitive nonlinear operations and involve densely connected layers for classification. However, studies in neurophysiology have revealed that EEG signals carry information in different ranges of frequency components. To better reflect these multi-frequency properties in EEGs, we propose a novel deep multi-scale neural network that discovers feature representations in multiple frequency/time ranges and extracts relationships among electrodes, i.e., spatial representations, for subject intention/condition identification. Furthermore, by completely representing EEG signals with spatio-spectral-temporal information, the proposed method can be utilized for diverse paradigms in both active and passive BCIs, contrary to existing methods that are primarily focused on single-paradigm BCIs. To demonstrate the validity of our proposed method, we conducted experiments on various paradigms of active/passive BCI datasets. Our experimental results demonstrated that the proposed method achieved performance improvements when judged against comparable state-of-the-art methods. Additionally, we analyzed the proposed method using different techniques, such as PSD curves and relevance score inspection to validate the multi-scale EEG signal information capturing ability, activation pattern maps for investigating the learned spatial filters, and t-SNE plotting for visualizing represented features. Finally, we also demonstrated our method's application to real-world problems.
Toward Subject Invariant and Class Disentangled Representation in BCI via Cross-Domain Mutual Information Estimator
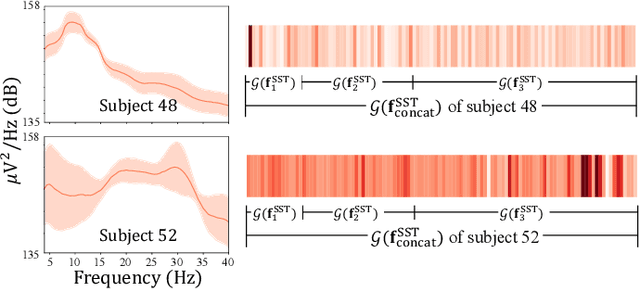
Oct 18, 2019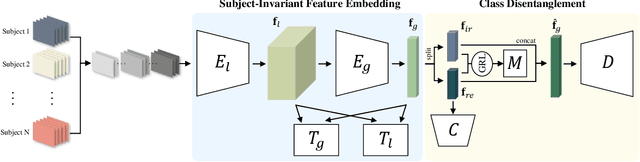
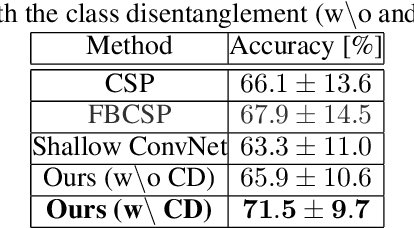
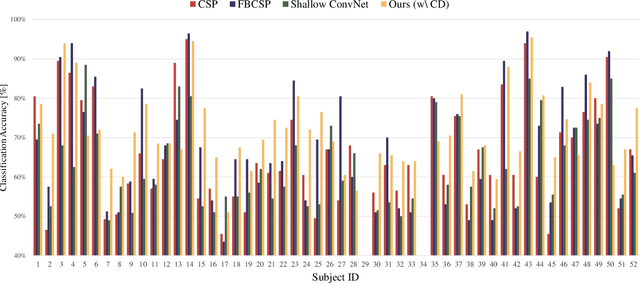

Abstract:In recent, deep learning-based feature representation methods have shown a promising impact in electroencephalography (EEG)-based brain-computer interface (BCI). Nonetheless, due to high intra- and inter-subject variabilities, many studies on decoding EEG were designed in a subject-specific manner by using calibration samples, with no much concern on its less practical, time-consuming, and data-hungry process. To tackle this problem, recent studies took advantage of transfer learning, especially using domain adaptation techniques. However, there still remain two challenging limitations; i) most domain adaptation methods are designed for labeled source and unlabeled target domain whereas BCI tasks generally have multiple annotated domains. ii) most of the methods do not consider negatively transferable to disrupt generalization ability. In this paper, we propose a novel network architecture to tackle those limitations by estimating mutual information in high-level representation and low-level representation, separately. Specifically, our proposed method extracts domain-invariant and class-relevant features, thereby enhancing generalizability in classification across. It is also noteworthy that our method can be applicable to a new subject with a small amount of data via a fine-tuning, step only, reducing calibration time for practical uses. We validated our proposed method on a big motor imagery EEG dataset by showing promising results, compared to competing methods considered in our experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge