Kwanseok Oh
IdenBAT: Disentangled Representation Learning for Identity-Preserved Brain Age Transformation
Oct 22, 2024Abstract:Brain age transformation aims to convert reference brain images into synthesized images that accurately reflect the age-specific features of a target age group. The primary objective of this task is to modify only the age-related attributes of the reference image while preserving all other age-irrelevant attributes. However, achieving this goal poses substantial challenges due to the inherent entanglement of various image attributes within features extracted from a backbone encoder, resulting in simultaneous alterations during the image generation. To address this challenge, we propose a novel architecture that employs disentangled representation learning for identity-preserved brain age transformation called IdenBAT. This approach facilitates the decomposition of image features, ensuring the preservation of individual traits while selectively transforming age-related characteristics to match those of the target age group. Through comprehensive experiments conducted on both 2D and full-size 3D brain datasets, our method adeptly converts input images to target age while retaining individual characteristics accurately. Furthermore, our approach demonstrates superiority over existing state-of-the-art regarding performance fidelity.
FIESTA: Fourier-Based Semantic Augmentation with Uncertainty Guidance for Enhanced Domain Generalizability in Medical Image Segmentation
Jun 20, 2024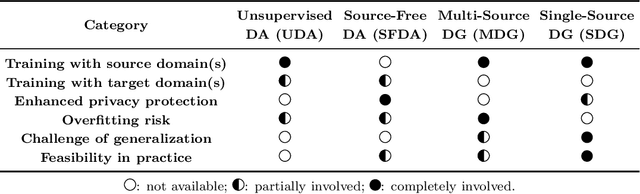
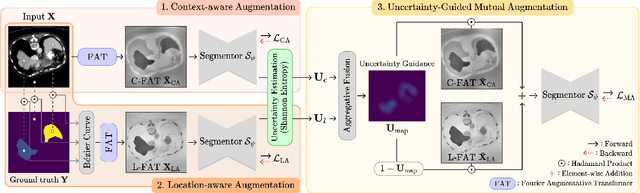
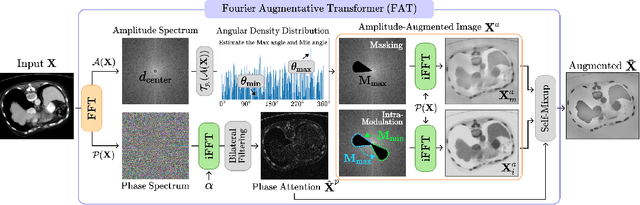
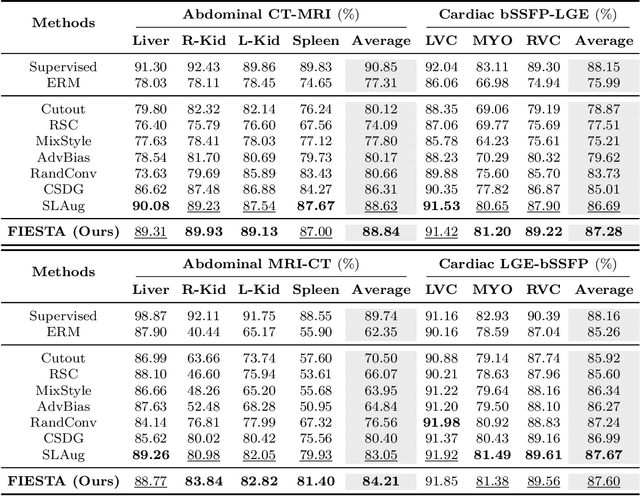
Abstract:Single-source domain generalization (SDG) in medical image segmentation (MIS) aims to generalize a model using data from only one source domain to segment data from an unseen target domain. Despite substantial advances in SDG with data augmentation, existing methods often fail to fully consider the details and uncertain areas prevalent in MIS, leading to mis-segmentation. This paper proposes a Fourier-based semantic augmentation method called FIESTA using uncertainty guidance to enhance the fundamental goals of MIS in an SDG context by manipulating the amplitude and phase components in the frequency domain. The proposed Fourier augmentative transformer addresses semantic amplitude modulation based on meaningful angular points to induce pertinent variations and harnesses the phase spectrum to ensure structural coherence. Moreover, FIESTA employs epistemic uncertainty to fine-tune the augmentation process, improving the ability of the model to adapt to diverse augmented data and concentrate on areas with higher ambiguity. Extensive experiments across three cross-domain scenarios demonstrate that FIESTA surpasses recent state-of-the-art SDG approaches in segmentation performance and significantly contributes to boosting the applicability of the model in medical imaging modalities.
Transferring Ultrahigh-Field Representations for Intensity-Guided Brain Segmentation of Low-Field Magnetic Resonance Imaging
Feb 13, 2024



Abstract:Ultrahigh-field (UHF) magnetic resonance imaging (MRI), i.e., 7T MRI, provides superior anatomical details of internal brain structures owing to its enhanced signal-to-noise ratio and susceptibility-induced contrast. However, the widespread use of 7T MRI is limited by its high cost and lower accessibility compared to low-field (LF) MRI. This study proposes a deep-learning framework that systematically fuses the input LF magnetic resonance feature representations with the inferred 7T-like feature representations for brain image segmentation tasks in a 7T-absent environment. Specifically, our adaptive fusion module aggregates 7T-like features derived from the LF image by a pre-trained network and then refines them to be effectively assimilable UHF guidance into LF image features. Using intensity-guided features obtained from such aggregation and assimilation, segmentation models can recognize subtle structural representations that are usually difficult to recognize when relying only on LF features. Beyond such advantages, this strategy can seamlessly be utilized by modulating the contrast of LF features in alignment with UHF guidance, even when employing arbitrary segmentation models. Exhaustive experiments demonstrated that the proposed method significantly outperformed all baseline models on both brain tissue and whole-brain segmentation tasks; further, it exhibited remarkable adaptability and scalability by successfully integrating diverse segmentation models and tasks. These improvements were not only quantifiable but also visible in the superlative visual quality of segmentation masks.
Domain Generalization for Medical Image Analysis: A Survey
Oct 05, 2023



Abstract:Medical Image Analysis (MedIA) has become an essential tool in medicine and healthcare, aiding in disease diagnosis, prognosis, and treatment planning, and recent successes in deep learning (DL) have made significant contributions to its advances. However, DL models for MedIA remain challenging to deploy in real-world situations, failing for generalization under the distributional gap between training and testing samples, known as a distribution shift problem. Researchers have dedicated their efforts to developing various DL methods to adapt and perform robustly on unknown and out-of-distribution data distributions. This paper comprehensively reviews domain generalization studies specifically tailored for MedIA. We provide a holistic view of how domain generalization techniques interact within the broader MedIA system, going beyond methodologies to consider the operational implications on the entire MedIA workflow. Specifically, we categorize domain generalization methods into data-level, feature-level, model-level, and analysis-level methods. We show how those methods can be used in various stages of the MedIA workflow with DL equipped from data acquisition to model prediction and analysis. Furthermore, we include benchmark datasets and applications used to evaluate these approaches and analyze the strengths and weaknesses of various methods, unveiling future research opportunities.
A Quantitatively Interpretable Model for Alzheimer's Disease Prediction Using Deep Counterfactuals
Oct 05, 2023



Abstract:Deep learning (DL) for predicting Alzheimer's disease (AD) has provided timely intervention in disease progression yet still demands attentive interpretability to explain how their DL models make definitive decisions. Recently, counterfactual reasoning has gained increasing attention in medical research because of its ability to provide a refined visual explanatory map. However, such visual explanatory maps based on visual inspection alone are insufficient unless we intuitively demonstrate their medical or neuroscientific validity via quantitative features. In this study, we synthesize the counterfactual-labeled structural MRIs using our proposed framework and transform it into a gray matter density map to measure its volumetric changes over the parcellated region of interest (ROI). We also devised a lightweight linear classifier to boost the effectiveness of constructed ROIs, promoted quantitative interpretation, and achieved comparable predictive performance to DL methods. Throughout this, our framework produces an ``AD-relatedness index'' for each ROI and offers an intuitive understanding of brain status for an individual patient and across patient groups with respect to AD progression.
XADLiME: eXplainable Alzheimer's Disease Likelihood Map Estimation via Clinically-guided Prototype Learning
Jul 27, 2022



Abstract:Diagnosing Alzheimer's disease (AD) involves a deliberate diagnostic process owing to its innate traits of irreversibility with subtle and gradual progression. These characteristics make AD biomarker identification from structural brain imaging (e.g., structural MRI) scans quite challenging. Furthermore, there is a high possibility of getting entangled with normal aging. We propose a novel deep-learning approach through eXplainable AD Likelihood Map Estimation (XADLiME) for AD progression modeling over 3D sMRIs using clinically-guided prototype learning. Specifically, we establish a set of topologically-aware prototypes onto the clusters of latent clinical features, uncovering an AD spectrum manifold. We then measure the similarities between latent clinical features and well-established prototypes, estimating a "pseudo" likelihood map. By considering this pseudo map as an enriched reference, we employ an estimating network to estimate the AD likelihood map over a 3D sMRI scan. Additionally, we promote the explainability of such a likelihood map by revealing a comprehensible overview from two perspectives: clinical and morphological. During the inference, this estimated likelihood map served as a substitute over unseen sMRI scans for effectively conducting the downstream task while providing thorough explainable states.
Learn-Explain-Reinforce: Counterfactual Reasoning and Its Guidance to Reinforce an Alzheimer's Disease Diagnosis Model
Aug 21, 2021



Abstract:Existing studies on disease diagnostic models focus either on diagnostic model learning for performance improvement or on the visual explanation of a trained diagnostic model. We propose a novel learn-explain-reinforce (LEAR) framework that unifies diagnostic model learning, visual explanation generation (explanation unit), and trained diagnostic model reinforcement (reinforcement unit) guided by the visual explanation. For the visual explanation, we generate a counterfactual map that transforms an input sample to be identified as an intended target label. For example, a counterfactual map can localize hypothetical abnormalities within a normal brain image that may cause it to be diagnosed with Alzheimer's disease (AD). We believe that the generated counterfactual maps represent data-driven and model-induced knowledge about a target task, i.e., AD diagnosis using structural MRI, which can be a vital source of information to reinforce the generalization of the trained diagnostic model. To this end, we devise an attention-based feature refinement module with the guidance of the counterfactual maps. The explanation and reinforcement units are reciprocal and can be operated iteratively. Our proposed approach was validated via qualitative and quantitative analysis on the ADNI dataset. Its comprehensibility and fidelity were demonstrated through ablation studies and comparisons with existing methods.
Born Identity Network: Multi-way Counterfactual Map Generation to Explain a Classifier's Decision
Nov 24, 2020
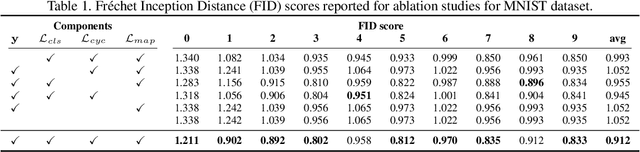
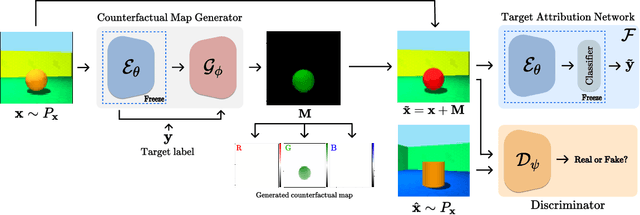
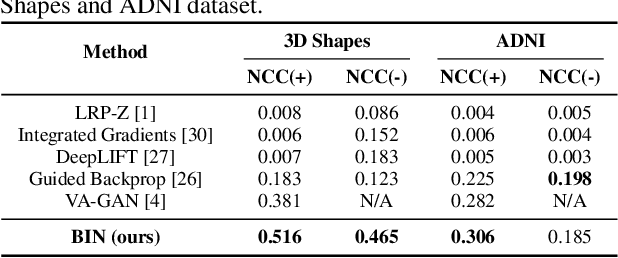
Abstract:There exists an apparent negative correlation between performance and interpretability of deep learning models. In an effort to reduce this negative correlation, we propose Born Identity Network (BIN), which is a post-hoc approach for producing multi-way counterfactual maps. A counterfactual map transforms an input sample to be classified as a target label, which is similar to how humans process knowledge through counterfactual thinking. Thus, producing a better counterfactual map may be a step towards explanation at the level of human knowledge. For example, a counterfactual map can localize hypothetical abnormalities from a normal brain image that may cause it to be diagnosed with a disease. Specifically, our proposed BIN consists of two core components: Counterfactual Map Generator and Target Attribution Network. The Counterfactual Map Generator is a variation of conditional GAN which can synthesize a counterfactual map conditioned on an arbitrary target label. The Target Attribution Network works in a complementary manner to enforce target label attribution to the synthesized map. We have validated our proposed BIN in qualitative, quantitative analysis on MNIST, 3D Shapes, and ADNI datasets, and show the comprehensibility and fidelity of our method from various ablation studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge