Jee Seok Yoon
Domain Generalization for Medical Image Analysis: A Survey
Oct 05, 2023



Abstract:Medical Image Analysis (MedIA) has become an essential tool in medicine and healthcare, aiding in disease diagnosis, prognosis, and treatment planning, and recent successes in deep learning (DL) have made significant contributions to its advances. However, DL models for MedIA remain challenging to deploy in real-world situations, failing for generalization under the distributional gap between training and testing samples, known as a distribution shift problem. Researchers have dedicated their efforts to developing various DL methods to adapt and perform robustly on unknown and out-of-distribution data distributions. This paper comprehensively reviews domain generalization studies specifically tailored for MedIA. We provide a holistic view of how domain generalization techniques interact within the broader MedIA system, going beyond methodologies to consider the operational implications on the entire MedIA workflow. Specifically, we categorize domain generalization methods into data-level, feature-level, model-level, and analysis-level methods. We show how those methods can be used in various stages of the MedIA workflow with DL equipped from data acquisition to model prediction and analysis. Furthermore, we include benchmark datasets and applications used to evaluate these approaches and analyze the strengths and weaknesses of various methods, unveiling future research opportunities.
SADM: Sequence-Aware Diffusion Model for Longitudinal Medical Image Generation
Dec 16, 2022Abstract:Human organs constantly undergo anatomical changes due to a complex mix of short-term (e.g., heartbeat) and long-term (e.g., aging) factors. Evidently, prior knowledge of these factors will be beneficial when modeling their future state, i.e., via image generation. However, most of the medical image generation tasks only rely on the input from a single image, thus ignoring the sequential dependency even when longitudinal data is available. Sequence-aware deep generative models, where model input is a sequence of ordered and timestamped images, are still underexplored in the medical imaging domain that is featured by several unique challenges: 1) Sequences with various lengths; 2) Missing data or frame, and 3) High dimensionality. To this end, we propose a sequence-aware diffusion model (SADM) for the generation of longitudinal medical images. Recently, diffusion models have shown promising results on high-fidelity image generation. Our method extends this new technique by introducing a sequence-aware transformer as the conditional module in a diffusion model. The novel design enables learning longitudinal dependency even with missing data during training and allows autoregressive generation of a sequence of images during inference. Our extensive experiments on 3D longitudinal medical images demonstrate the effectiveness of SADM compared with baselines and alternative methods.
XADLiME: eXplainable Alzheimer's Disease Likelihood Map Estimation via Clinically-guided Prototype Learning
Jul 27, 2022



Abstract:Diagnosing Alzheimer's disease (AD) involves a deliberate diagnostic process owing to its innate traits of irreversibility with subtle and gradual progression. These characteristics make AD biomarker identification from structural brain imaging (e.g., structural MRI) scans quite challenging. Furthermore, there is a high possibility of getting entangled with normal aging. We propose a novel deep-learning approach through eXplainable AD Likelihood Map Estimation (XADLiME) for AD progression modeling over 3D sMRIs using clinically-guided prototype learning. Specifically, we establish a set of topologically-aware prototypes onto the clusters of latent clinical features, uncovering an AD spectrum manifold. We then measure the similarities between latent clinical features and well-established prototypes, estimating a "pseudo" likelihood map. By considering this pseudo map as an enriched reference, we employ an estimating network to estimate the AD likelihood map over a 3D sMRI scan. Additionally, we promote the explainability of such a likelihood map by revealing a comprehensible overview from two perspectives: clinical and morphological. During the inference, this estimated likelihood map served as a substitute over unseen sMRI scans for effectively conducting the downstream task while providing thorough explainable states.
Learn-Explain-Reinforce: Counterfactual Reasoning and Its Guidance to Reinforce an Alzheimer's Disease Diagnosis Model
Aug 21, 2021



Abstract:Existing studies on disease diagnostic models focus either on diagnostic model learning for performance improvement or on the visual explanation of a trained diagnostic model. We propose a novel learn-explain-reinforce (LEAR) framework that unifies diagnostic model learning, visual explanation generation (explanation unit), and trained diagnostic model reinforcement (reinforcement unit) guided by the visual explanation. For the visual explanation, we generate a counterfactual map that transforms an input sample to be identified as an intended target label. For example, a counterfactual map can localize hypothetical abnormalities within a normal brain image that may cause it to be diagnosed with Alzheimer's disease (AD). We believe that the generated counterfactual maps represent data-driven and model-induced knowledge about a target task, i.e., AD diagnosis using structural MRI, which can be a vital source of information to reinforce the generalization of the trained diagnostic model. To this end, we devise an attention-based feature refinement module with the guidance of the counterfactual maps. The explanation and reinforcement units are reciprocal and can be operated iteratively. Our proposed approach was validated via qualitative and quantitative analysis on the ADNI dataset. Its comprehensibility and fidelity were demonstrated through ablation studies and comparisons with existing methods.
Born Identity Network: Multi-way Counterfactual Map Generation to Explain a Classifier's Decision
Nov 24, 2020
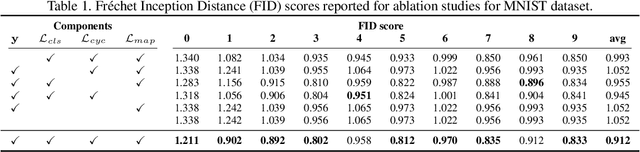
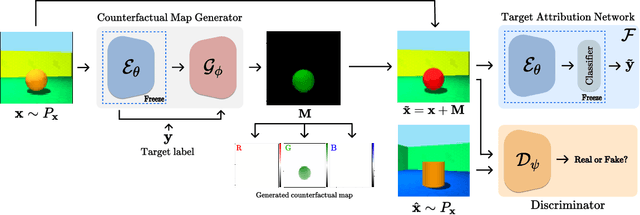
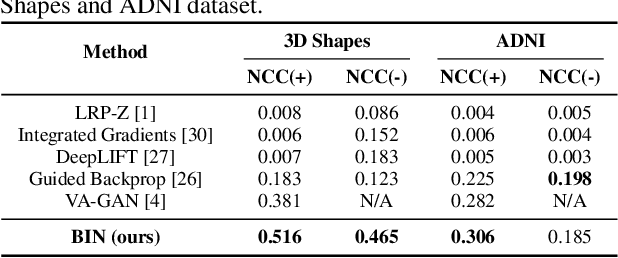
Abstract:There exists an apparent negative correlation between performance and interpretability of deep learning models. In an effort to reduce this negative correlation, we propose Born Identity Network (BIN), which is a post-hoc approach for producing multi-way counterfactual maps. A counterfactual map transforms an input sample to be classified as a target label, which is similar to how humans process knowledge through counterfactual thinking. Thus, producing a better counterfactual map may be a step towards explanation at the level of human knowledge. For example, a counterfactual map can localize hypothetical abnormalities from a normal brain image that may cause it to be diagnosed with a disease. Specifically, our proposed BIN consists of two core components: Counterfactual Map Generator and Target Attribution Network. The Counterfactual Map Generator is a variation of conditional GAN which can synthesize a counterfactual map conditioned on an arbitrary target label. The Target Attribution Network works in a complementary manner to enforce target label attribution to the synthesized map. We have validated our proposed BIN in qualitative, quantitative analysis on MNIST, 3D Shapes, and ADNI datasets, and show the comprehensibility and fidelity of our method from various ablation studies.
Toward Subject Invariant and Class Disentangled Representation in BCI via Cross-Domain Mutual Information Estimator
Oct 18, 2019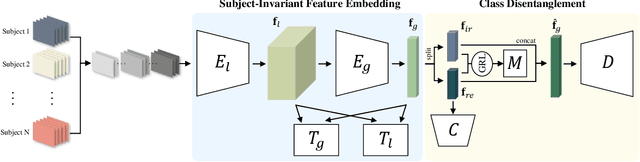
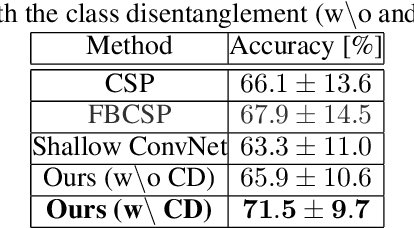
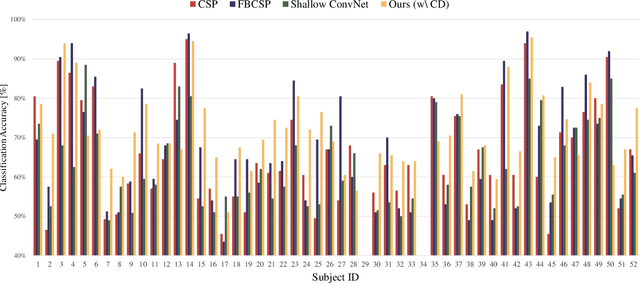

Abstract:In recent, deep learning-based feature representation methods have shown a promising impact in electroencephalography (EEG)-based brain-computer interface (BCI). Nonetheless, due to high intra- and inter-subject variabilities, many studies on decoding EEG were designed in a subject-specific manner by using calibration samples, with no much concern on its less practical, time-consuming, and data-hungry process. To tackle this problem, recent studies took advantage of transfer learning, especially using domain adaptation techniques. However, there still remain two challenging limitations; i) most domain adaptation methods are designed for labeled source and unlabeled target domain whereas BCI tasks generally have multiple annotated domains. ii) most of the methods do not consider negatively transferable to disrupt generalization ability. In this paper, we propose a novel network architecture to tackle those limitations by estimating mutual information in high-level representation and low-level representation, separately. Specifically, our proposed method extracts domain-invariant and class-relevant features, thereby enhancing generalizability in classification across. It is also noteworthy that our method can be applicable to a new subject with a small amount of data via a fine-tuning, step only, reducing calibration time for practical uses. We validated our proposed method on a big motor imagery EEG dataset by showing promising results, compared to competing methods considered in our experiments.
Plug-in Factorization for Latent Representation Disentanglement
May 29, 2019



Abstract:In this work, we propose a Factorized Disentangler-Entangler Network (FDEN) that learns to decompose a latent representation into two mutually independent factors, namely, identity and style. Given a latent representation, the proposed framework draws a set of interpretable factors aligned to identity of an observed data and learns to maximize the independency between these factors. Our work introduces an idea for a plug-in method to disentangle latent representations of already learned deep models with no affect to the model. In doing so, it brings the possibilities of extending state-of-the-art models to solve different tasks and also maintain the performance of its original task. Thus, FDEN is naturally applicable to jointly perform multiple tasks such as few-shot learning and image-to-image translation in a single framework. We show the effectiveness of our work in disentangling a latent representation in two parts. First, to evaluate the alignment of factor to an identity, we perform few-shot learning using only the aligned factor. Then, to evaluate the effectiveness of decomposition of latent representation and to show that plugin method does not affect the deep model in its performance, we perform image-to-image style transfer by mixing factors of different images. These evaluations show, qualitatively and quantitatively, that our proposed framework can indeed disentangle a latent representation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge