Wentao Cui
Comprehensive Metapath-based Heterogeneous Graph Transformer for Gene-Disease Association Prediction
Jan 14, 2025



Abstract:Discovering gene-disease associations is crucial for understanding disease mechanisms, yet identifying these associations remains challenging due to the time and cost of biological experiments. Computational methods are increasingly vital for efficient and scalable gene-disease association prediction. Graph-based learning models, which leverage node features and network relationships, are commonly employed for biomolecular predictions. However, existing methods often struggle to effectively integrate node features, heterogeneous structures, and semantic information. To address these challenges, we propose COmprehensive MEtapath-based heterogeneous graph Transformer(COMET) for predicting gene-disease associations. COMET integrates diverse datasets to construct comprehensive heterogeneous networks, initializing node features with BioGPT. We define seven Metapaths and utilize a transformer framework to aggregate Metapath instances, capturing global contexts and long-distance dependencies. Through intra- and inter-metapath aggregation using attention mechanisms, COMET fuses latent vectors from multiple Metapaths to enhance GDA prediction accuracy. Our method demonstrates superior robustness compared to state-of-the-art approaches. Ablation studies and visualizations validate COMET's effectiveness, providing valuable insights for advancing human health research.
Towards Graph Prompt Learning: A Survey and Beyond
Aug 26, 2024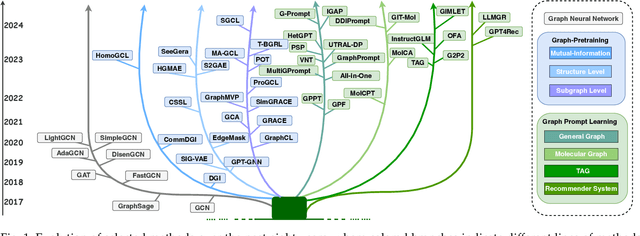
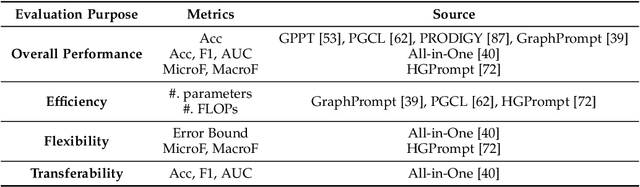
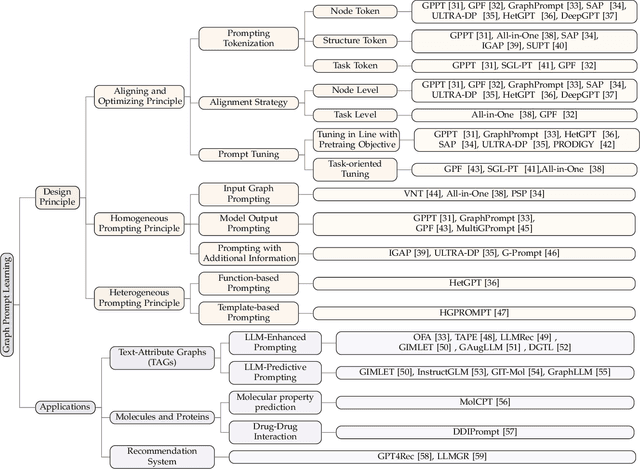
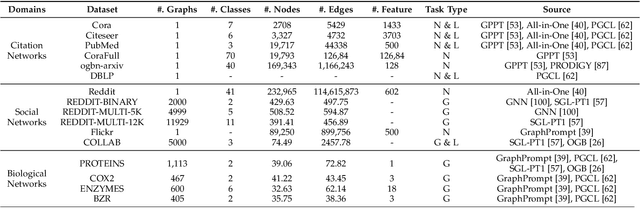
Abstract:Large-scale "pre-train and prompt learning" paradigms have demonstrated remarkable adaptability, enabling broad applications across diverse domains such as question answering, image recognition, and multimodal retrieval. This approach fully leverages the potential of large-scale pre-trained models, reducing downstream data requirements and computational costs while enhancing model applicability across various tasks. Graphs, as versatile data structures that capture relationships between entities, play pivotal roles in fields such as social network analysis, recommender systems, and biological graphs. Despite the success of pre-train and prompt learning paradigms in Natural Language Processing (NLP) and Computer Vision (CV), their application in graph domains remains nascent. In graph-structured data, not only do the node and edge features often have disparate distributions, but the topological structures also differ significantly. This diversity in graph data can lead to incompatible patterns or gaps between pre-training and fine-tuning on downstream graphs. We aim to bridge this gap by summarizing methods for alleviating these disparities. This includes exploring prompt design methodologies, comparing related techniques, assessing application scenarios and datasets, and identifying unresolved problems and challenges. This survey categorizes over 100 relevant works in this field, summarizing general design principles and the latest applications, including text-attributed graphs, molecules, proteins, and recommendation systems. Through this extensive review, we provide a foundational understanding of graph prompt learning, aiming to impact not only the graph mining community but also the broader Artificial General Intelligence (AGI) community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge