Vladimir Gligorijević
Implicitly Guided Design with PropEn: Match your Data to Follow the Gradient
May 28, 2024Abstract:Across scientific domains, generating new models or optimizing existing ones while meeting specific criteria is crucial. Traditional machine learning frameworks for guided design use a generative model and a surrogate model (discriminator), requiring large datasets. However, real-world scientific applications often have limited data and complex landscapes, making data-hungry models inefficient or impractical. We propose a new framework, PropEn, inspired by ``matching'', which enables implicit guidance without training a discriminator. By matching each sample with a similar one that has a better property value, we create a larger training dataset that inherently indicates the direction of improvement. Matching, combined with an encoder-decoder architecture, forms a domain-agnostic generative framework for property enhancement. We show that training with a matched dataset approximates the gradient of the property of interest while remaining within the data distribution, allowing efficient design optimization. Extensive evaluations in toy problems and scientific applications, such as therapeutic protein design and airfoil optimization, demonstrate PropEn's advantages over common baselines. Notably, the protein design results are validated with wet lab experiments, confirming the competitiveness and effectiveness of our approach.
A Pareto-optimal compositional energy-based model for sampling and optimization of protein sequences
Oct 19, 2022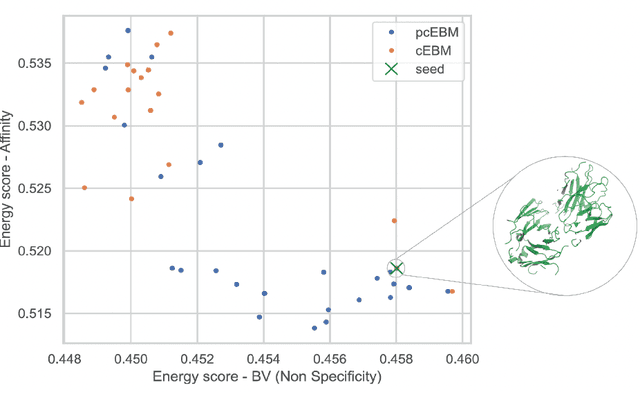
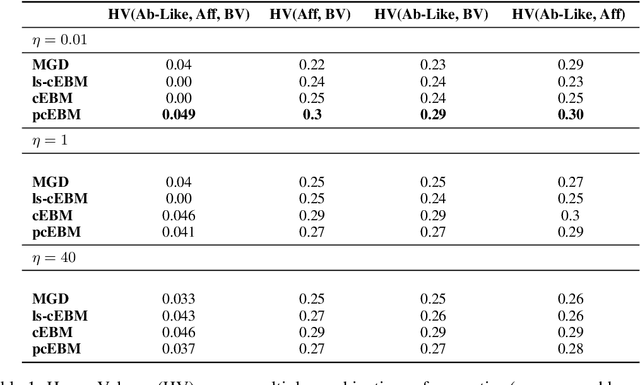
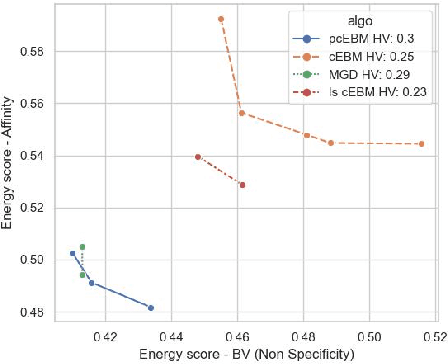
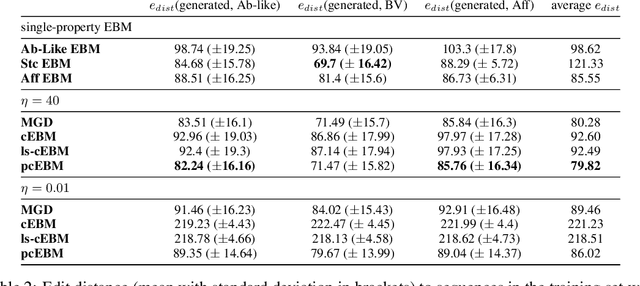
Abstract:Deep generative models have emerged as a popular machine learning-based approach for inverse design problems in the life sciences. However, these problems often require sampling new designs that satisfy multiple properties of interest in addition to learning the data distribution. This multi-objective optimization becomes more challenging when properties are independent or orthogonal to each other. In this work, we propose a Pareto-compositional energy-based model (pcEBM), a framework that uses multiple gradient descent for sampling new designs that adhere to various constraints in optimizing distinct properties. We demonstrate its ability to learn non-convex Pareto fronts and generate sequences that simultaneously satisfy multiple desired properties across a series of real-world antibody design tasks.
Multi-segment preserving sampling for deep manifold sampler
May 09, 2022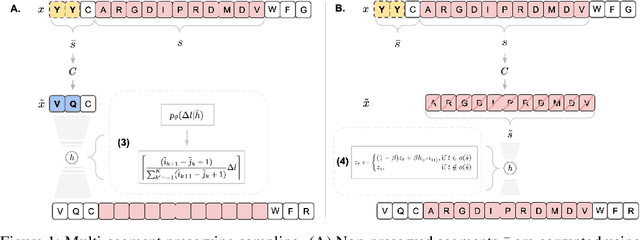
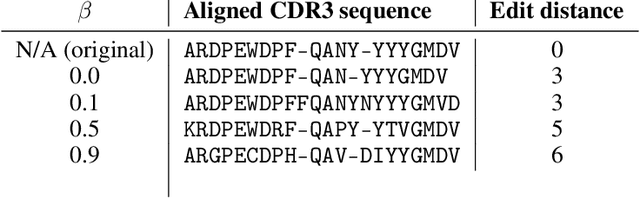
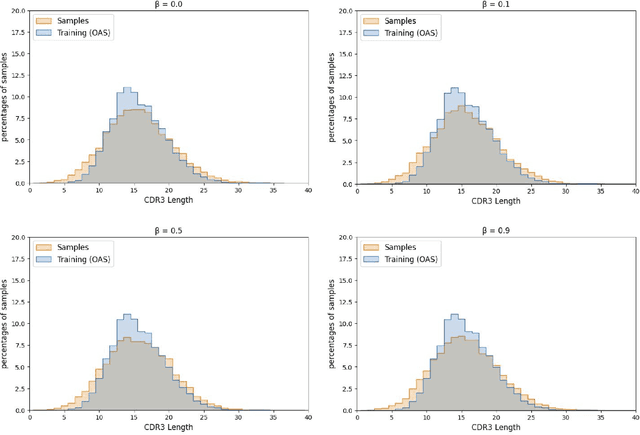
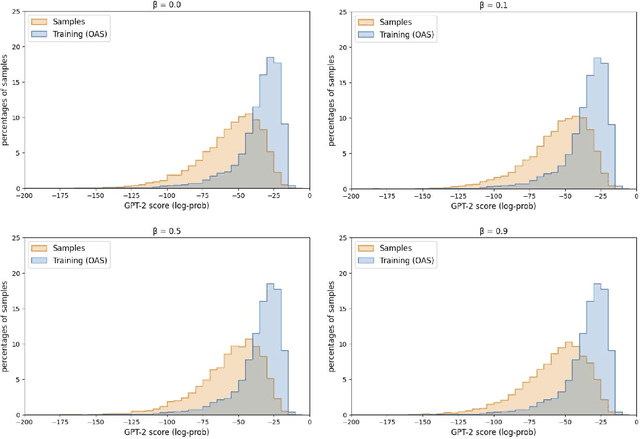
Abstract:Deep generative modeling for biological sequences presents a unique challenge in reconciling the bias-variance trade-off between explicit biological insight and model flexibility. The deep manifold sampler was recently proposed as a means to iteratively sample variable-length protein sequences by exploiting the gradients from a function predictor. We introduce an alternative approach to this guided sampling procedure, multi-segment preserving sampling, that enables the direct inclusion of domain-specific knowledge by designating preserved and non-preserved segments along the input sequence, thereby restricting variation to only select regions. We present its effectiveness in the context of antibody design by training two models: a deep manifold sampler and a GPT-2 language model on nearly six million heavy chain sequences annotated with the IGHV1-18 gene. During sampling, we restrict variation to only the complementarity-determining region 3 (CDR3) of the input. We obtain log probability scores from a GPT-2 model for each sampled CDR3 and demonstrate that multi-segment preserving sampling generates reasonable designs while maintaining the desired, preserved regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge