Tian Zhao
Medical Multimodal Foundation Models in Clinical Diagnosis and Treatment: Applications, Challenges, and Future Directions
Dec 03, 2024



Abstract:Recent advancements in deep learning have significantly revolutionized the field of clinical diagnosis and treatment, offering novel approaches to improve diagnostic precision and treatment efficacy across diverse clinical domains, thus driving the pursuit of precision medicine. The growing availability of multi-organ and multimodal datasets has accelerated the development of large-scale Medical Multimodal Foundation Models (MMFMs). These models, known for their strong generalization capabilities and rich representational power, are increasingly being adapted to address a wide range of clinical tasks, from early diagnosis to personalized treatment strategies. This review offers a comprehensive analysis of recent developments in MMFMs, focusing on three key aspects: datasets, model architectures, and clinical applications. We also explore the challenges and opportunities in optimizing multimodal representations and discuss how these advancements are shaping the future of healthcare by enabling improved patient outcomes and more efficient clinical workflows.
GCtx-UNet: Efficient Network for Medical Image Segmentation
Jun 09, 2024Abstract:Medical image segmentation is crucial for disease diagnosis and monitoring. Though effective, the current segmentation networks such as UNet struggle with capturing long-range features. More accurate models such as TransUNet, Swin-UNet, and CS-UNet have higher computation complexity. To address this problem, we propose GCtx-UNet, a lightweight segmentation architecture that can capture global and local image features with accuracy better or comparable to the state-of-the-art approaches. GCtx-UNet uses vision transformer that leverages global context self-attention modules joined with local self-attention to model long and short range spatial dependencies. GCtx-UNet is evaluated on the Synapse multi-organ abdominal CT dataset, the ACDC cardiac MRI dataset, and several polyp segmentation datasets. In terms of Dice Similarity Coefficient (DSC) and Hausdorff Distance (HD) metrics, GCtx-UNet outperformed CNN-based and Transformer-based approaches, with notable gains in the segmentation of complex and small anatomical structures. Moreover, GCtx-UNet is much more efficient than the state-of-the-art approaches with smaller model size, lower computation workload, and faster training and inference speed, making it a practical choice for clinical applications.
Transfer Learning for Microstructure Segmentation with CS-UNet: A Hybrid Algorithm with Transformer and CNN Encoders
Aug 26, 2023Abstract:Transfer learning improves the performance of deep learning models by initializing them with parameters pre-trained on larger datasets. Intuitively, transfer learning is more effective when pre-training is on the in-domain datasets. A recent study by NASA has demonstrated that the microstructure segmentation with encoder-decoder algorithms benefits more from CNN encoders pre-trained on microscopy images than from those pre-trained on natural images. However, CNN models only capture the local spatial relations in images. In recent years, attention networks such as Transformers are increasingly used in image analysis to capture the long-range relations between pixels. In this study, we compare the segmentation performance of Transformer and CNN models pre-trained on microscopy images with those pre-trained on natural images. Our result partially confirms the NASA study that the segmentation performance of out-of-distribution images (taken under different imaging and sample conditions) is significantly improved when pre-training on microscopy images. However, the performance gain for one-shot and few-shot learning is more modest with Transformers. We also find that for image segmentation, the combination of pre-trained Transformers and CNN encoders are consistently better than pre-trained CNN encoders alone. Our dataset (of about 50,000 images) combines the public portion of the NASA dataset with additional images we collected. Even with much less training data, our pre-trained models have significantly better performance for image segmentation. This result suggests that Transformers and CNN complement each other and when pre-trained on microscopy images, they are more beneficial to the downstream tasks.
Computer Vision Methods for the Microstructural Analysis of Materials: The State-of-the-art and Future Perspectives
Jul 29, 2022Abstract:Finding quantitative descriptors representing the microstructural features of a given material is an ongoing research area in the paradigm of Materials-by-Design. Historically, microstructural analysis mostly relies on qualitative descriptions. However, to build a robust and accurate process-structure-properties relationship, which is required for designing new advanced high-performance materials, the extraction of quantitative and meaningful statistical data from the microstructural analysis is a critical step. In recent years, computer vision (CV) methods, especially those which are centered around convolutional neural network (CNN) algorithms have shown promising results for this purpose. This review paper focuses on the state-of-the-art CNN-based techniques that have been applied to various multi-scale microstructural image analysis tasks, including classification, object detection, segmentation, feature extraction, and reconstruction. Additionally, we identified the main challenges with regard to the application of these methods to materials science research. Finally, we discussed some possible future directions of research in this area. In particular, we emphasized the application of transformer-based models and their capabilities to improve the microstructural analysis of materials.
Efficient Memory Partitioning in Software Defined Hardware
Feb 02, 2022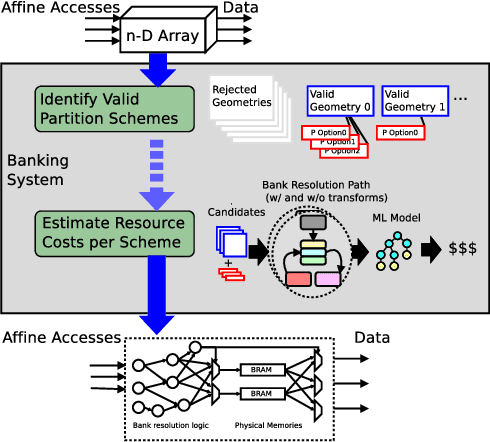
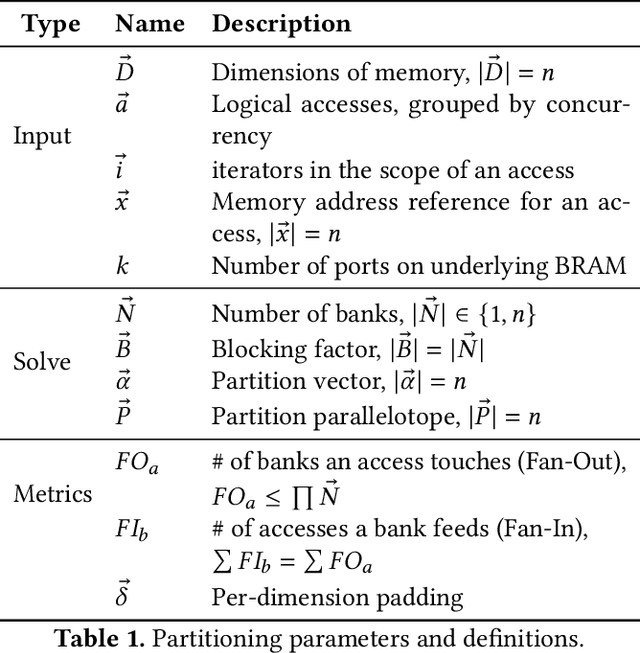
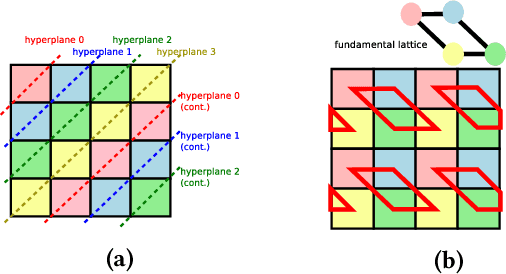
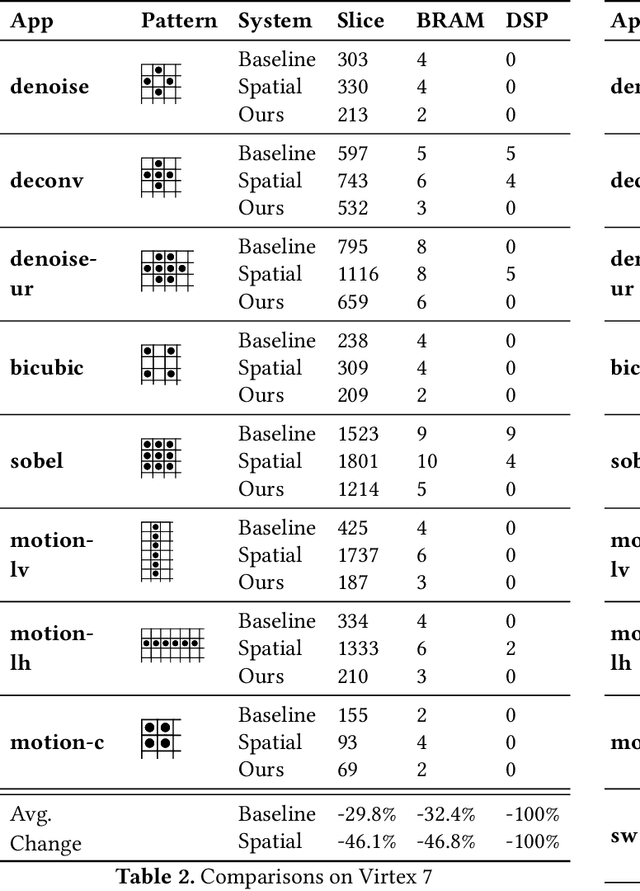
Abstract:As programmers turn to software-defined hardware (SDH) to maintain a high level of productivity while programming hardware to run complex algorithms, heavy-lifting must be done by the compiler to automatically partition on-chip arrays. In this paper, we introduce an automatic memory partitioning system that can quickly compute more efficient partitioning schemes than prior systems. Our system employs a variety of resource-saving optimizations and an ML cost model to select the best partitioning scheme from an array of candidates. We compared our system against various state-of-the-art SDH compilers and FPGAs on a variety of benchmarks and found that our system generates solutions that, on average, consume 40.3% fewer logic resources, 78.3% fewer FFs, 54.9% fewer Block RAMs (BRAMs), and 100% fewer DSPs.
Serving Recurrent Neural Networks Efficiently with a Spatial Accelerator
Sep 26, 2019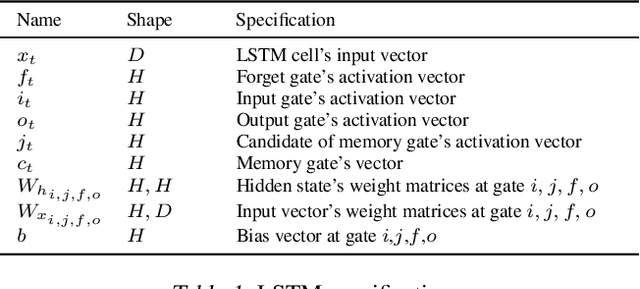
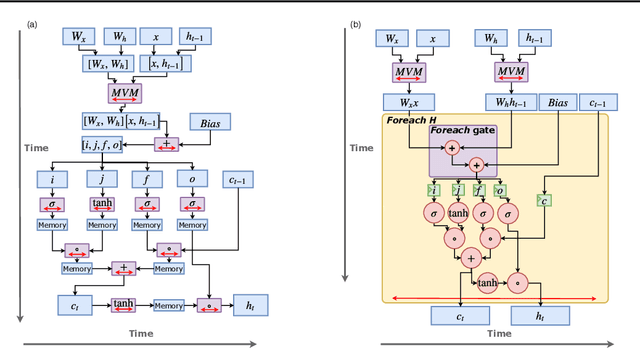
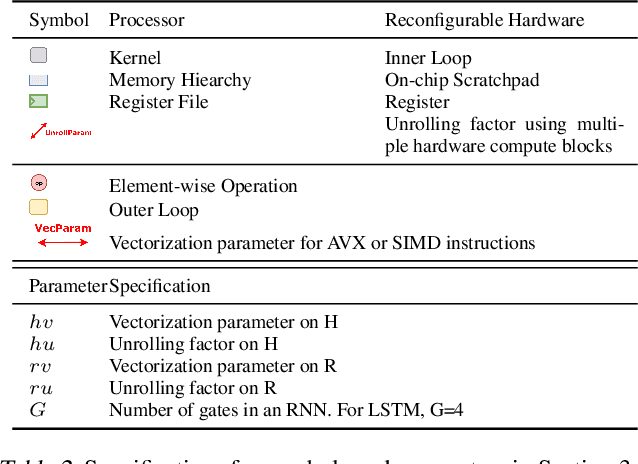
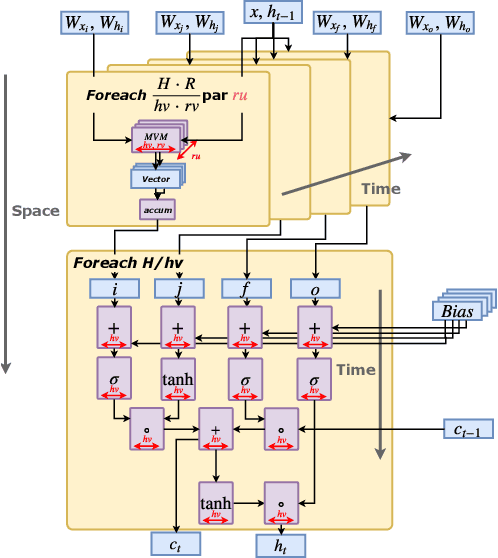
Abstract:Recurrent Neural Network (RNN) applications form a major class of AI-powered, low-latency data center workloads. Most execution models for RNN acceleration break computation graphs into BLAS kernels, which lead to significant inter-kernel data movement and resource underutilization. We show that by supporting more general loop constructs that capture design parameters in accelerators, it is possible to improve resource utilization using cross-kernel optimization without sacrificing programmability. Such abstraction level enables a design space search that can lead to efficient usage of on-chip resources on a spatial architecture across a range of problem sizes. We evaluate our optimization strategy on such abstraction with DeepBench using a configurable spatial accelerator. We demonstrate that this implementation provides a geometric speedup of 30x in performance, 1.6x in area, and 2x in power efficiency compared to a Tesla V100 GPU, and a geometric speedup of 2x compared to Microsoft Brainwave implementation on a Stratix 10 FPGA.
Analysis of DAWNBench, a Time-to-Accuracy Machine Learning Performance Benchmark
Jun 04, 2018

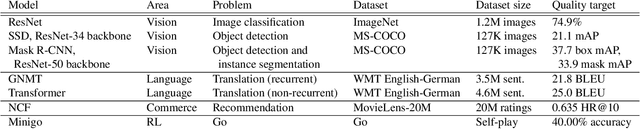
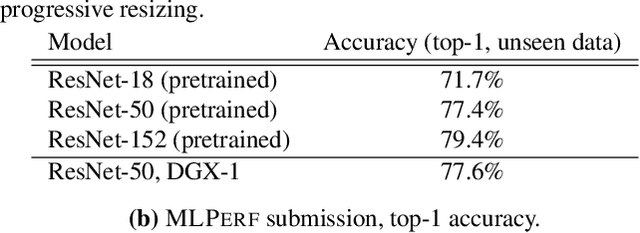
Abstract:The deep learning community has proposed optimizations spanning hardware, software, and learning theory to improve the computational performance of deep learning workloads. While some of these optimizations perform the same operations faster (e.g., switching from a NVIDIA K80 to P100), many modify the semantics of the training procedure (e.g., large minibatch training, reduced precision), which can impact a model's generalization ability. Due to a lack of standard evaluation criteria that considers these trade-offs, it has become increasingly difficult to compare these different advances. To address this shortcoming, DAWNBENCH and the upcoming MLPERF benchmarks use time-to-accuracy as the primary metric for evaluation, with the accuracy threshold set close to state-of-the-art and measured on a held-out dataset not used in training; the goal is to train to this accuracy threshold as fast as possible. In DAWNBENCH , the winning entries improved time-to-accuracy on ImageNet by two orders of magnitude over the seed entries. Despite this progress, it is unclear how sensitive time-to-accuracy is to the chosen threshold as well as the variance between independent training runs, and how well models optimized for time-to-accuracy generalize. In this paper, we provide evidence to suggest that time-to-accuracy has a low coefficient of variance and that the models tuned for it generalize nearly as well as pre-trained models. We additionally analyze the winning entries to understand the source of these speedups, and give recommendations for future benchmarking efforts.
DeepDSL: A Compilation-based Domain-Specific Language for Deep Learning
Jan 09, 2017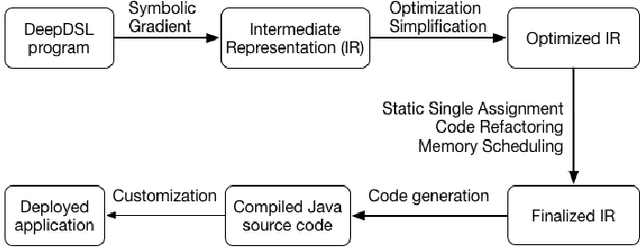

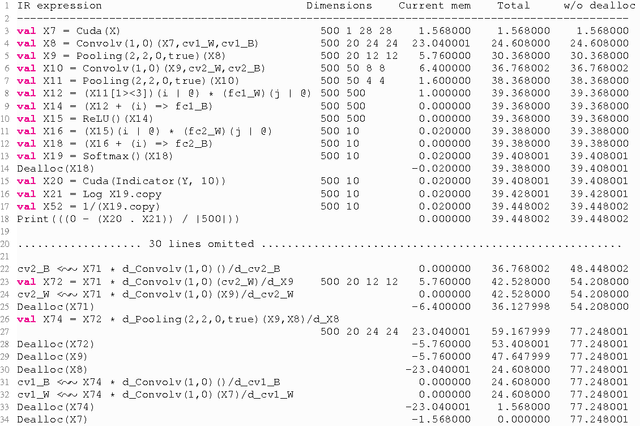
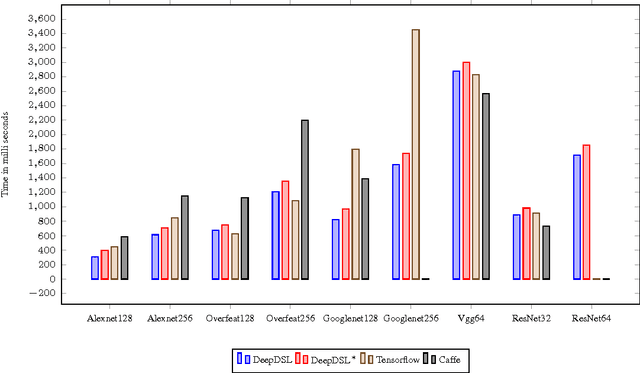
Abstract:In recent years, Deep Learning (DL) has found great success in domains such as multimedia understanding. However, the complex nature of multimedia data makes it difficult to develop DL-based software. The state-of-the art tools, such as Caffe, TensorFlow, Torch7, and CNTK, while are successful in their applicable domains, are programming libraries with fixed user interface, internal representation, and execution environment. This makes it difficult to implement portable and customized DL applications. In this paper, we present DeepDSL, a domain specific language (DSL) embedded in Scala, that compiles deep networks written in DeepDSL to Java source code. Deep DSL provides (1) intuitive constructs to support compact encoding of deep networks; (2) symbolic gradient derivation of the networks; (3) static analysis for memory consumption and error detection; and (4) DSL-level optimization to improve memory and runtime efficiency. DeepDSL programs are compiled into compact, efficient, customizable, and portable Java source code, which operates the CUDA and CUDNN interfaces running on Nvidia GPU via a Java Native Interface (JNI) library. We evaluated DeepDSL with a number of popular DL networks. Our experiments show that the compiled programs have very competitive runtime performance and memory efficiency compared to the existing libraries.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge