Steve Farrell
Lawrence Berkeley National Laboratory
MatterChat: A Multi-Modal LLM for Material Science
Feb 18, 2025


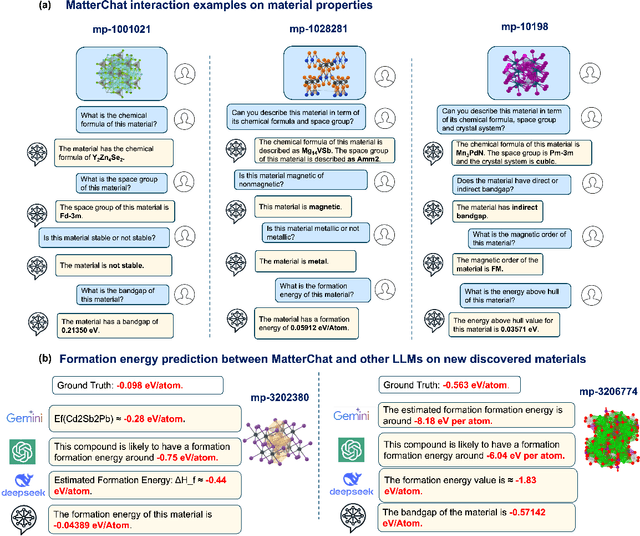
Abstract:Understanding and predicting the properties of inorganic materials is crucial for accelerating advancements in materials science and driving applications in energy, electronics, and beyond. Integrating material structure data with language-based information through multi-modal large language models (LLMs) offers great potential to support these efforts by enhancing human-AI interaction. However, a key challenge lies in integrating atomic structures at full resolution into LLMs. In this work, we introduce MatterChat, a versatile structure-aware multi-modal LLM that unifies material structural data and textual inputs into a single cohesive model. MatterChat employs a bridging module to effectively align a pretrained machine learning interatomic potential with a pretrained LLM, reducing training costs and enhancing flexibility. Our results demonstrate that MatterChat significantly improves performance in material property prediction and human-AI interaction, surpassing general-purpose LLMs such as GPT-4. We also demonstrate its usefulness in applications such as more advanced scientific reasoning and step-by-step material synthesis.
Physics and Computing Performance of the Exa.TrkX TrackML Pipeline
Mar 11, 2021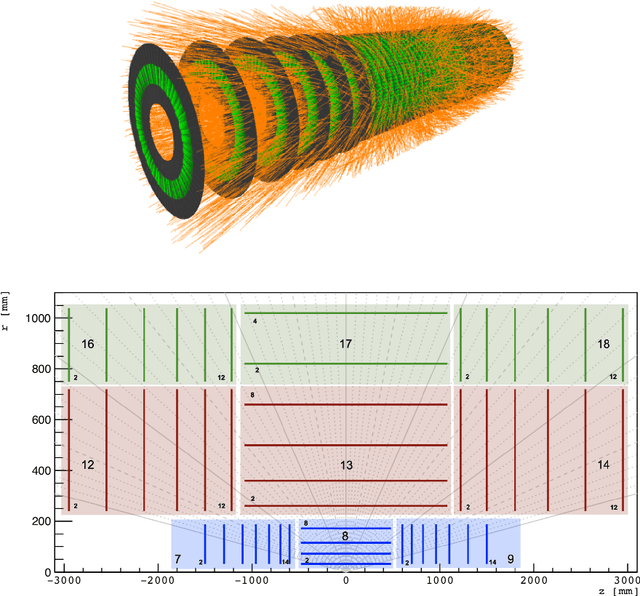
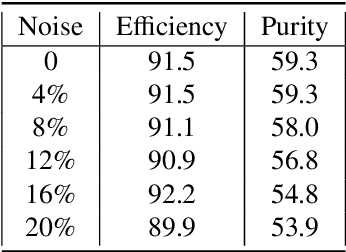
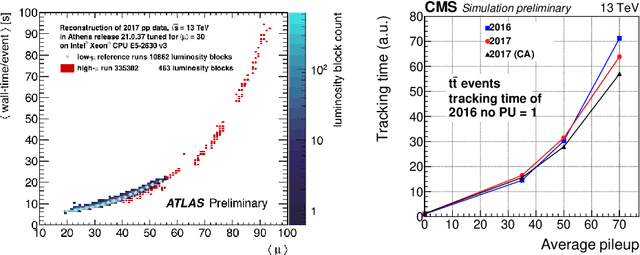
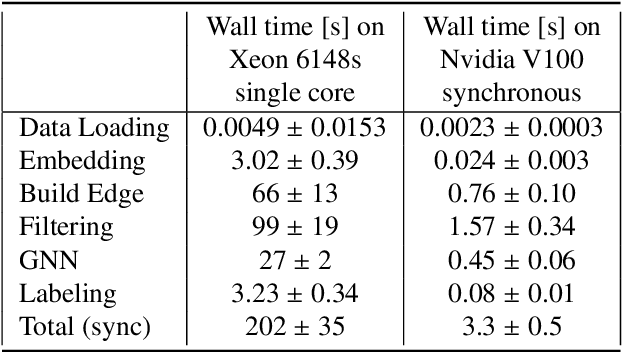
Abstract:The Exa.TrkX project has applied geometric learning concepts such as metric learning and graph neural networks to HEP particle tracking. The Exa.TrkX tracking pipeline clusters detector measurements to form track candidates and filters them. The pipeline, originally developed using the TrackML dataset (a simulation of an LHC-like tracking detector), has been demonstrated on various detectors, including the DUNE LArTPC and the CMS High-Granularity Calorimeter. This paper documents new developments needed to study the physics and computing performance of the Exa.TrkX pipeline on the full TrackML dataset, a first step towards validating the pipeline using ATLAS and CMS data. The pipeline achieves tracking efficiency and purity similar to production tracking algorithms. Crucially for future HEP applications, the pipeline benefits significantly from GPU acceleration, and its computational requirements scale close to linearly with the number of particles in the event.
A Molecular-MNIST Dataset for Machine Learning Study on Diffraction Imaging and Microscopy
Nov 15, 2019


Abstract:An image dataset of 10 different size molecules, where each molecule has 2,000 structural variants, is generated from the 2D cross-sectional projection of Molecular Dynamics trajectories. The purpose of this dataset is to provide a benchmark dataset for the increasing need of machine learning, deep learning and image processing on the study of scattering, imaging and microscopy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge