Stefano D'Aronco
Zero-Shot Bird Species Recognition by Learning from Field Guides
Jun 03, 2022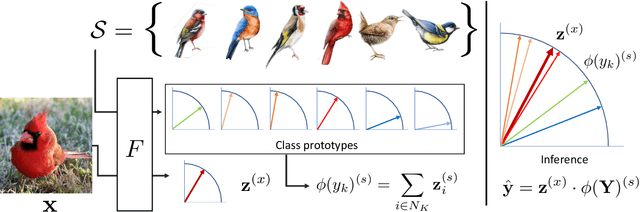
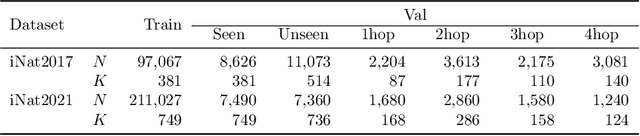
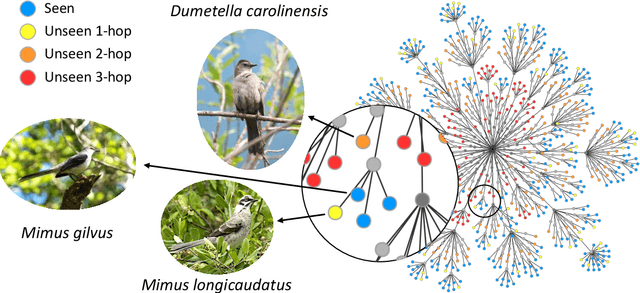

Abstract:We exploit field guides to learn bird species recognition, in particular zero-shot recognition of unseen species. The illustrations contained in field guides deliberately focus on discriminative properties of a species, and can serve as side information to transfer knowledge from seen to unseen classes. We study two approaches: (1) a contrastive encoding of illustrations that can be fed into zero-shot learning schemes; and (2) a novel method that leverages the fact that illustrations are also images and as such structurally more similar to photographs than other kinds of side information. Our results show that illustrations from field guides, which are readily available for a wide range of species, are indeed a competitive source of side information. On the iNaturalist2021 subset, we obtain a harmonic mean from 749 seen and 739 unseen classes greater than $45\%$ (@top-10) and $15\%$ (@top-1). Which shows that field guides are a valuable option for challenging real-world scenarios with many species.
FiLM-Ensemble: Probabilistic Deep Learning via Feature-wise Linear Modulation
May 31, 2022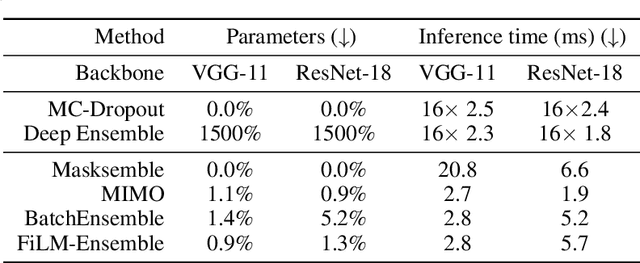
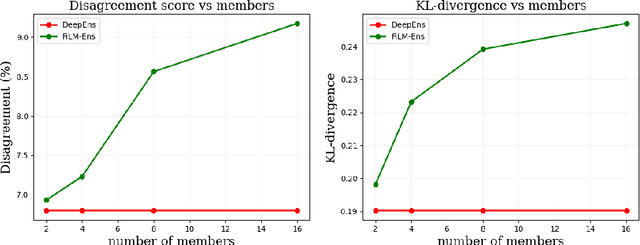
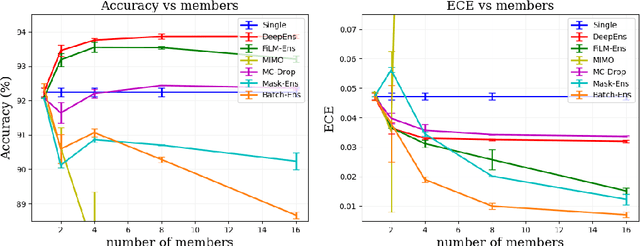
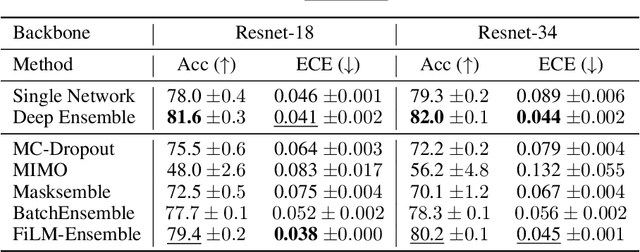
Abstract:The ability to estimate epistemic uncertainty is often crucial when deploying machine learning in the real world, but modern methods often produce overconfident, uncalibrated uncertainty predictions. A common approach to quantify epistemic uncertainty, usable across a wide class of prediction models, is to train a model ensemble. In a naive implementation, the ensemble approach has high computational cost and high memory demand. This challenges in particular modern deep learning, where even a single deep network is already demanding in terms of compute and memory, and has given rise to a number of attempts to emulate the model ensemble without actually instantiating separate ensemble members. We introduce FiLM-Ensemble, a deep, implicit ensemble method based on the concept of Feature-wise Linear Modulation (FiLM). That technique was originally developed for multi-task learning, with the aim of decoupling different tasks. We show that the idea can be extended to uncertainty quantification: by modulating the network activations of a single deep network with FiLM, one obtains a model ensemble with high diversity, and consequently well-calibrated estimates of epistemic uncertainty, with low computational overhead in comparison. Empirically, FiLM-Ensemble outperforms other implicit ensemble methods, and it and comes very close to the upper bound of an explicit ensemble of networks (sometimes even beating it), at a fraction of the memory cost.
Learning Graph Regularisation for Guided Super-Resolution
Mar 27, 2022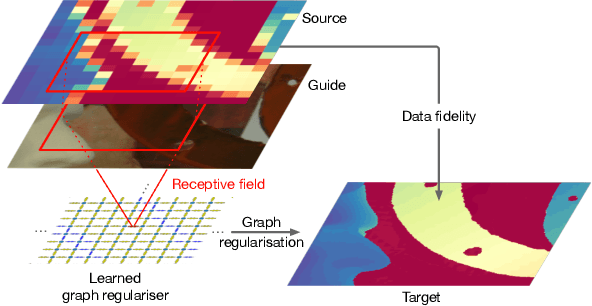

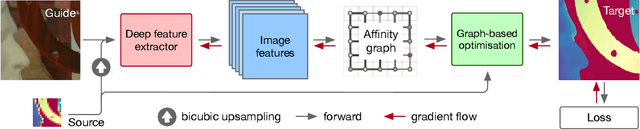

Abstract:We introduce a novel formulation for guided super-resolution. Its core is a differentiable optimisation layer that operates on a learned affinity graph. The learned graph potentials make it possible to leverage rich contextual information from the guide image, while the explicit graph optimisation within the architecture guarantees rigorous fidelity of the high-resolution target to the low-resolution source. With the decision to employ the source as a constraint rather than only as an input to the prediction, our method differs from state-of-the-art deep architectures for guided super-resolution, which produce targets that, when downsampled, will only approximately reproduce the source. This is not only theoretically appealing, but also produces crisper, more natural-looking images. A key property of our method is that, although the graph connectivity is restricted to the pixel lattice, the associated edge potentials are learned with a deep feature extractor and can encode rich context information over large receptive fields. By taking advantage of the sparse graph connectivity, it becomes possible to propagate gradients through the optimisation layer and learn the edge potentials from data. We extensively evaluate our method on several datasets, and consistently outperform recent baselines in terms of quantitative reconstruction errors, while also delivering visually sharper outputs. Moreover, we demonstrate that our method generalises particularly well to new datasets not seen during training.
A Deep Learning Approach for Digital ColorReconstruction of Lenticular Films
Feb 10, 2022



Abstract:We propose the first accurate digitization and color reconstruction process for historical lenticular film that is robust to artifacts. Lenticular films emerged in the 1920s and were one of the first technologies that permitted to capture full color information in motion. The technology leverages an RGB filter and cylindrical lenticules embossed on the film surface to encode the color in the horizontal spatial dimension of the image. To project the pictures the encoding process was reversed using an appropriate analog device. In this work, we introduce an automated, fully digital pipeline to process the scan of lenticular films and colorize the image. Our method merges deep learning with a model-based approach in order to maximize the performance while making sure that the reconstructed colored images truthfully match the encoded color information. Our model employs different strategies to achieve an effective color reconstruction, in particular (i) we use data augmentation to create a robust lenticule segmentation network, (ii) we fit the lenticules raster prediction to obtain a precise vectorial lenticule localization, and (iii) we train a colorization network that predicts interpolation coefficients in order to obtain a truthful colorization. We validate the proposed method on a lenticular film dataset and compare it to other approaches. Since no colored groundtruth is available as reference, we conduct a user study to validate our method in a subjective manner. The results of the study show that the proposed method is largely preferred with respect to other existing and baseline methods.
Digital Taxonomist: Identifying Plant Species in Citizen Scientists' Photographs
Jun 07, 2021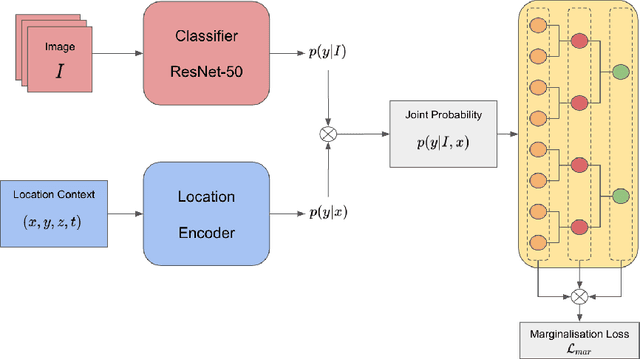
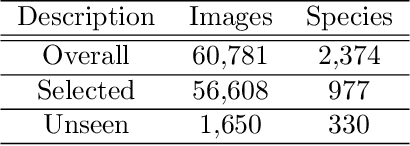
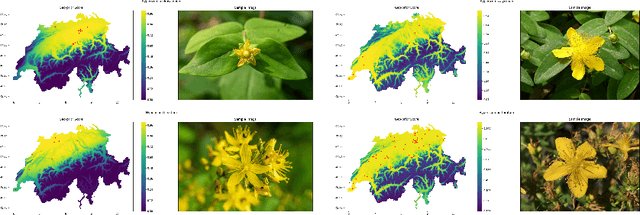
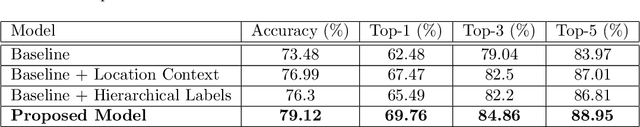
Abstract:Automatic identification of plant specimens from amateur photographs could improve species range maps, thus supporting ecosystems research as well as conservation efforts. However, classifying plant specimens based on image data alone is challenging: some species exhibit large variations in visual appearance, while at the same time different species are often visually similar; additionally, species observations follow a highly imbalanced, long-tailed distribution due to differences in abundance as well as observer biases. On the other hand, most species observations are accompanied by side information about the spatial, temporal and ecological context. Moreover, biological species are not an unordered list of classes but embedded in a hierarchical taxonomic structure. We propose a machine learning model that takes into account these additional cues in a unified framework. Our Digital Taxonomist is able to identify plant species in photographs more correctly.
Mapping oil palm density at country scale: An active learning approach
May 24, 2021



Abstract:Accurate mapping of oil palm is important for understanding its past and future impact on the environment. We propose to map and count oil palms by estimating tree densities per pixel for large-scale analysis. This allows for fine-grained analysis, for example regarding different planting patterns. To that end, we propose a new, active deep learning method to estimate oil palm density at large scale from Sentinel-2 satellite images, and apply it to generate complete maps for Malaysia and Indonesia. What makes the regression of oil palm density challenging is the need for representative reference data that covers all relevant geographical conditions across a large territory. Specifically for density estimation, generating reference data involves counting individual trees. To keep the associated labelling effort low we propose an active learning (AL) approach that automatically chooses the most relevant samples to be labelled. Our method relies on estimates of the epistemic model uncertainty and of the diversity among samples, making it possible to retrieve an entire batch of relevant samples in a single iteration. Moreover, our algorithm has linear computational complexity and is easily parallelisable to cover large areas. We use our method to compute the first oil palm density map with $10\,$m Ground Sampling Distance (GSD) , for all of Indonesia and Malaysia and for two different years, 2017 and 2019. The maps have a mean absolute error of $\pm$7.3 trees/$ha$, estimated from an independent validation set. We also analyse density variations between different states within a country and compare them to official estimates. According to our estimates there are, in total, $>1.2$ billion oil palms in Indonesia covering $>$15 million $ha$, and $>0.5$ billion oil palms in Malaysia covering $>6$ million $ha$.
PC2WF: 3D Wireframe Reconstruction from Raw Point Clouds
Mar 04, 2021



Abstract:We introduce PC2WF, the first end-to-end trainable deep network architecture to convert a 3D point cloud into a wireframe model. The network takes as input an unordered set of 3D points sampled from the surface of some object, and outputs a wireframe of that object, i.e., a sparse set of corner points linked by line segments. Recovering the wireframe is a challenging task, where the numbers of both vertices and edges are different for every instance, and a-priori unknown. Our architecture gradually builds up the model: It starts by encoding the points into feature vectors. Based on those features, it identifies a pool of candidate vertices, then prunes those candidates to a final set of corner vertices and refines their locations. Next, the corners are linked with an exhaustive set of candidate edges, which is again pruned to obtain the final wireframe. All steps are trainable, and errors can be backpropagated through the entire sequence. We validate the proposed model on a publicly available synthetic dataset, for which the ground truth wireframes are accessible, as well as on a new real-world dataset. Our model produces wireframe abstractions of good quality and outperforms several baselines.
Crop mapping from image time series: deep learning with multi-scale label hierarchies
Feb 17, 2021



Abstract:The aim of this paper is to map agricultural crops by classifying satellite image time series. Domain experts in agriculture work with crop type labels that are organised in a hierarchical tree structure, where coarse classes (like orchards) are subdivided into finer ones (like apples, pears, vines, etc.). We develop a crop classification method that exploits this expert knowledge and significantly improves the mapping of rare crop types. The three-level label hierarchy is encoded in a convolutional, recurrent neural network (convRNN), such that for each pixel the model predicts three labels at different level of granularity. This end-to-end trainable, hierarchical network architecture allows the model to learn joint feature representations of rare classes (e.g., apples, pears) at a coarser level (e.g., orchard), thereby boosting classification performance at the fine-grained level. Additionally, labelling at different granularity also makes it possible to adjust the output according to the classification scores; as coarser labels with high confidence are sometimes more useful for agricultural practice than fine-grained but very uncertain labels. We validate the proposed method on a new, large dataset that we make public. ZueriCrop covers an area of 50 km x 48 km in the Swiss cantons of Zurich and Thurgau with a total of 116'000 individual fields spanning 48 crop classes, and 28,000 (multi-temporal) image patches from Sentinel-2. We compare our proposed hierarchical convRNN model with several baselines, including methods designed for imbalanced class distributions. The hierarchical approach performs superior by at least 9.9 percentage points in F1-score.
Crop Classification under Varying Cloud Cover with Neural Ordinary Differential Equations
Dec 04, 2020



Abstract:Optical satellite sensors cannot see the Earth's surface through clouds. Despite the periodic revisit cycle, image sequences acquired by Earth observation satellites are therefore irregularly sampled in time. State-of-the-art methods for crop classification (and other time series analysis tasks) rely on techniques that implicitly assume regular temporal spacing between observations, such as recurrent neural networks (RNNs). We propose to use neural ordinary differential equations (NODEs) in combination with RNNs to classify crop types in irregularly spaced image sequences. The resulting ODE-RNN models consist of two steps: an update step, where a recurrent unit assimilates new input data into the model's hidden state; and a prediction step, in which NODE propagates the hidden state until the next observation arrives. The prediction step is based on a continuous representation of the latent dynamics, which has several advantages. At the conceptual level, it is a more natural way to describe the mechanisms that govern the phenological cycle. From a practical point of view, it makes it possible to sample the system state at arbitrary points in time, such that one can integrate observations whenever they are available, and extrapolate beyond the last observation. Our experiments show that ODE-RNN indeed improves classification accuracy over common baselines such as LSTM, GRU, and temporal convolution. The gains are most prominent in the challenging scenario where only few observations are available (i.e., frequent cloud cover). Moreover, we show that the ability to extrapolate translates to better classification performance early in the season, which is important for forecasting.
Deep Active Learning in Remote Sensing for data efficient Change Detection
Aug 25, 2020



Abstract:We investigate active learning in the context of deep neural network models for change detection and map updating. Active learning is a natural choice for a number of remote sensing tasks, including the detection of local surface changes: changes are on the one hand rare and on the other hand their appearance is varied and diffuse, making it hard to collect a representative training set in advance. In the active learning setting, one starts from a minimal set of training examples and progressively chooses informative samples that are annotated by a user and added to the training set. Hence, a core component of an active learning system is a mechanism to estimate model uncertainty, which is then used to pick uncertain, informative samples. We study different mechanisms to capture and quantify this uncertainty when working with deep networks, based on the variance or entropy across explicit or implicit model ensembles. We show that active learning successfully finds highly informative samples and automatically balances the training distribution, and reaches the same performance as a model supervised with a large, pre-annotated training set, with $\approx$99% fewer annotated samples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge