Soroush Mehraban
CARE-PD: A Multi-Site Anonymized Clinical Dataset for Parkinson's Disease Gait Assessment
Oct 05, 2025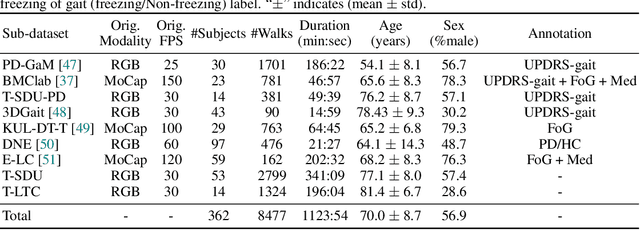
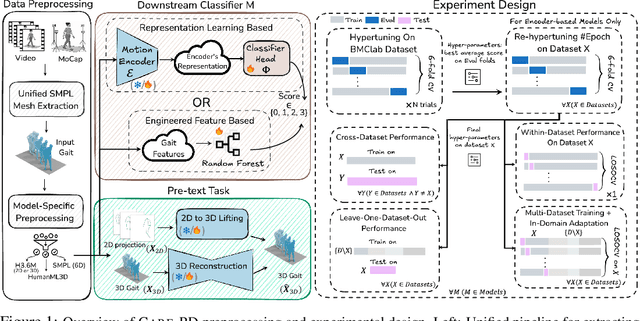
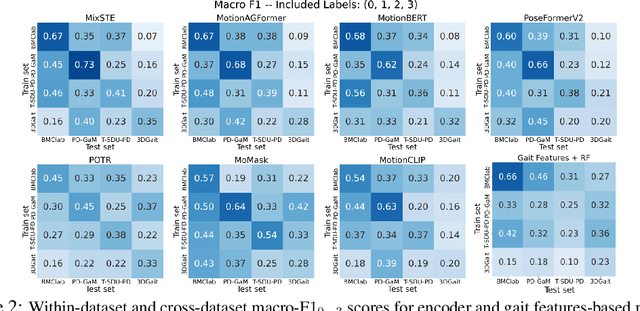
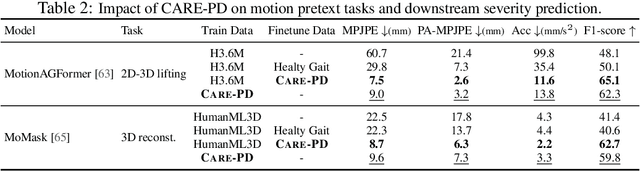
Abstract:Objective gait assessment in Parkinson's Disease (PD) is limited by the absence of large, diverse, and clinically annotated motion datasets. We introduce CARE-PD, the largest publicly available archive of 3D mesh gait data for PD, and the first multi-site collection spanning 9 cohorts from 8 clinical centers. All recordings (RGB video or motion capture) are converted into anonymized SMPL meshes via a harmonized preprocessing pipeline. CARE-PD supports two key benchmarks: supervised clinical score prediction (estimating Unified Parkinson's Disease Rating Scale, UPDRS, gait scores) and unsupervised motion pretext tasks (2D-to-3D keypoint lifting and full-body 3D reconstruction). Clinical prediction is evaluated under four generalization protocols: within-dataset, cross-dataset, leave-one-dataset-out, and multi-dataset in-domain adaptation. To assess clinical relevance, we compare state-of-the-art motion encoders with a traditional gait-feature baseline, finding that encoders consistently outperform handcrafted features. Pretraining on CARE-PD reduces MPJPE (from 60.8mm to 7.5mm) and boosts PD severity macro-F1 by 17 percentage points, underscoring the value of clinically curated, diverse training data. CARE-PD and all benchmark code are released for non-commercial research at https://neurips2025.care-pd.ca/.
Token Perturbation Guidance for Diffusion Models
Jun 10, 2025Abstract:Classifier-free guidance (CFG) has become an essential component of modern diffusion models to enhance both generation quality and alignment with input conditions. However, CFG requires specific training procedures and is limited to conditional generation. To address these limitations, we propose Token Perturbation Guidance (TPG), a novel method that applies perturbation matrices directly to intermediate token representations within the diffusion network. TPG employs a norm-preserving shuffling operation to provide effective and stable guidance signals that improve generation quality without architectural changes. As a result, TPG is training-free and agnostic to input conditions, making it readily applicable to both conditional and unconditional generation. We further analyze the guidance term provided by TPG and show that its effect on sampling more closely resembles CFG compared to existing training-free guidance techniques. Extensive experiments on SDXL and Stable Diffusion 2.1 show that TPG achieves nearly a 2$\times$ improvement in FID for unconditional generation over the SDXL baseline, while closely matching CFG in prompt alignment. These results establish TPG as a general, condition-agnostic guidance method that brings CFG-like benefits to a broader class of diffusion models. The code is available at https://github.com/TaatiTeam/Token-Perturbation-Guidance
GAITGen: Disentangled Motion-Pathology Impaired Gait Generative Model -- Bringing Motion Generation to the Clinical Domain
Mar 28, 2025Abstract:Gait analysis is crucial for the diagnosis and monitoring of movement disorders like Parkinson's Disease. While computer vision models have shown potential for objectively evaluating parkinsonian gait, their effectiveness is limited by scarce clinical datasets and the challenge of collecting large and well-labelled data, impacting model accuracy and risk of bias. To address these gaps, we propose GAITGen, a novel framework that generates realistic gait sequences conditioned on specified pathology severity levels. GAITGen employs a Conditional Residual Vector Quantized Variational Autoencoder to learn disentangled representations of motion dynamics and pathology-specific factors, coupled with Mask and Residual Transformers for conditioned sequence generation. GAITGen generates realistic, diverse gait sequences across severity levels, enriching datasets and enabling large-scale model training in parkinsonian gait analysis. Experiments on our new PD-GaM (real) dataset demonstrate that GAITGen outperforms adapted state-of-the-art models in both reconstruction fidelity and generation quality, accurately capturing critical pathology-specific gait features. A clinical user study confirms the realism and clinical relevance of our generated sequences. Moreover, incorporating GAITGen-generated data into downstream tasks improves parkinsonian gait severity estimation, highlighting its potential for advancing clinical gait analysis.
LIFT: Latent Implicit Functions for Task- and Data-Agnostic Encoding
Mar 19, 2025Abstract:Implicit Neural Representations (INRs) are proving to be a powerful paradigm in unifying task modeling across diverse data domains, offering key advantages such as memory efficiency and resolution independence. Conventional deep learning models are typically modality-dependent, often requiring custom architectures and objectives for different types of signals. However, existing INR frameworks frequently rely on global latent vectors or exhibit computational inefficiencies that limit their broader applicability. We introduce LIFT, a novel, high-performance framework that addresses these challenges by capturing multiscale information through meta-learning. LIFT leverages multiple parallel localized implicit functions alongside a hierarchical latent generator to produce unified latent representations that span local, intermediate, and global features. This architecture facilitates smooth transitions across local regions, enhancing expressivity while maintaining inference efficiency. Additionally, we introduce ReLIFT, an enhanced variant of LIFT that incorporates residual connections and expressive frequency encodings. With this straightforward approach, ReLIFT effectively addresses the convergence-capacity gap found in comparable methods, providing an efficient yet powerful solution to improve capacity and speed up convergence. Empirical results show that LIFT achieves state-of-the-art (SOTA) performance in generative modeling and classification tasks, with notable reductions in computational costs. Moreover, in single-task settings, the streamlined ReLIFT architecture proves effective in signal representations and inverse problem tasks.
STARS: Self-supervised Tuning for 3D Action Recognition in Skeleton Sequences
Jul 15, 2024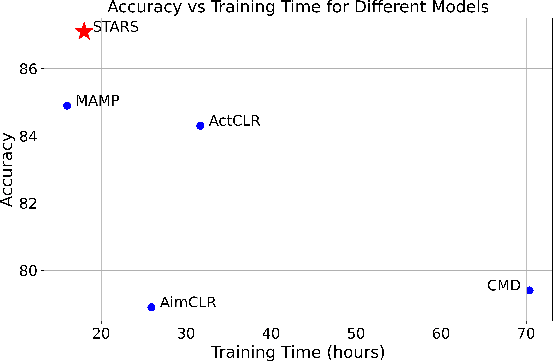
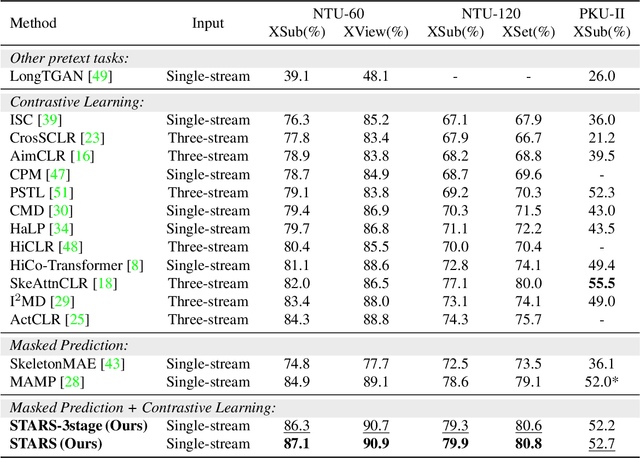
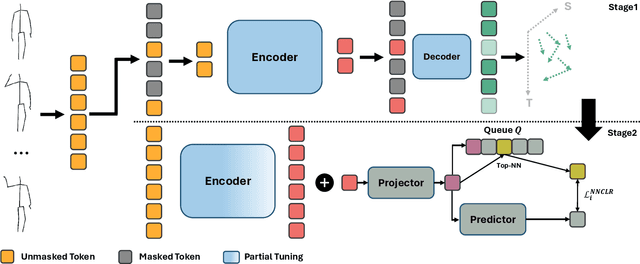
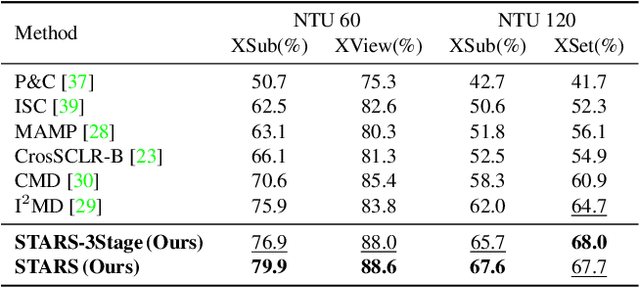
Abstract:Self-supervised pretraining methods with masked prediction demonstrate remarkable within-dataset performance in skeleton-based action recognition. However, we show that, unlike contrastive learning approaches, they do not produce well-separated clusters. Additionally, these methods struggle with generalization in few-shot settings. To address these issues, we propose Self-supervised Tuning for 3D Action Recognition in Skeleton sequences (STARS). Specifically, STARS first uses a masked prediction stage using an encoder-decoder architecture. It then employs nearest-neighbor contrastive learning to partially tune the weights of the encoder, enhancing the formation of semantic clusters for different actions. By tuning the encoder for a few epochs, and without using hand-crafted data augmentations, STARS achieves state-of-the-art self-supervised results in various benchmarks, including NTU-60, NTU-120, and PKU-MMD. In addition, STARS exhibits significantly better results than masked prediction models in few-shot settings, where the model has not seen the actions throughout pretraining. Project page: https://soroushmehraban.github.io/stars/
Benchmarking Skeleton-based Motion Encoder Models for Clinical Applications: Estimating Parkinson's Disease Severity in Walking Sequences
May 28, 2024



Abstract:This study investigates the application of general human motion encoders trained on large-scale human motion datasets for analyzing gait patterns in PD patients. Although these models have learned a wealth of human biomechanical knowledge, their effectiveness in analyzing pathological movements, such as parkinsonian gait, has yet to be fully validated. We propose a comparative framework and evaluate six pre-trained state-of-the-art human motion encoder models on their ability to predict the Movement Disorder Society - Unified Parkinson's Disease Rating Scale (MDS-UPDRS-III) gait scores from motion capture data. We compare these against a traditional gait feature-based predictive model in a recently released large public PD dataset, including PD patients on and off medication. The feature-based model currently shows higher weighted average accuracy, precision, recall, and F1-score. Motion encoder models with closely comparable results demonstrate promise for scalability and efficiency in clinical settings. This potential is underscored by the enhanced performance of the encoder model upon fine-tuning on PD training set. Four of the six human motion models examined provided prediction scores that were significantly different between on- and off-medication states. This finding reveals the sensitivity of motion encoder models to nuanced clinical changes. It also underscores the necessity for continued customization of these models to better capture disease-specific features, thereby reducing the reliance on labor-intensive feature engineering. Lastly, we establish a benchmark for the analysis of skeleton-based motion encoder models in clinical settings. To the best of our knowledge, this is the first study to provide a benchmark that enables state-of-the-art models to be tested and compete in a clinical context. Codes and benchmark leaderboard are available at code.
MotionAGFormer: Enhancing 3D Human Pose Estimation with a Transformer-GCNFormer Network
Oct 25, 2023



Abstract:Recent transformer-based approaches have demonstrated excellent performance in 3D human pose estimation. However, they have a holistic view and by encoding global relationships between all the joints, they do not capture the local dependencies precisely. In this paper, we present a novel Attention-GCNFormer (AGFormer) block that divides the number of channels by using two parallel transformer and GCNFormer streams. Our proposed GCNFormer module exploits the local relationship between adjacent joints, outputting a new representation that is complementary to the transformer output. By fusing these two representation in an adaptive way, AGFormer exhibits the ability to better learn the underlying 3D structure. By stacking multiple AGFormer blocks, we propose MotionAGFormer in four different variants, which can be chosen based on the speed-accuracy trade-off. We evaluate our model on two popular benchmark datasets: Human3.6M and MPI-INF-3DHP. MotionAGFormer-B achieves state-of-the-art results, with P1 errors of 38.4mm and 16.2mm, respectively. Remarkably, it uses a quarter of the parameters and is three times more computationally efficient than the previous leading model on Human3.6M dataset. Code and models are available at https://github.com/TaatiTeam/MotionAGFormer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge